A three-dimensional topology of complex I inferred from evolutionary correlations
- PMID: 22857522
- PMCID: PMC3436739
- DOI: 10.1186/1472-6807-12-19
A three-dimensional topology of complex I inferred from evolutionary correlations
Abstract
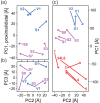
Background: The quaternary structure of eukaryotic NADH:ubiquinone oxidoreductase (complex I), the largest complex of the oxidative phosphorylation, is still mostly unresolved. Furthermore, it is unknown where transiently bound assembly factors interact with complex I. We therefore asked whether the evolution of complex I contains information about its 3D topology and the binding positions of its assembly factors. We approached these questions by correlating the evolutionary rates of eukaryotic complex I subunits using the mirror-tree method and mapping the results into a 3D representation by multidimensional scaling.
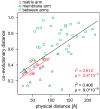
Results: More than 60% of the evolutionary correlation among the conserved seven subunits of the complex I matrix arm can be explained by the physical distance between the subunits. The three-dimensional evolutionary model of the eukaryotic conserved matrix arm has a striking similarity to the matrix arm quaternary structure in the bacterium Thermus thermophilus (rmsd=19 Å) and supports the previous finding that in eukaryotes the N-module is turned relative to the Q-module when compared to bacteria. By contrast, the evolutionary rates contained little information about the structure of the membrane arm. A large evolutionary model of 45 subunits and assembly factors allows to predict subunit positions and interactions (rmsd=52.6 Å). The model supports an interaction of NDUFAF3, C8orf38 and C2orf56 during the assembly of the proximal matrix arm and the membrane arm. The model further suggests a tight relationship between the assembly factor NUBPL and NDUFA2, which both have been linked to iron-sulfur cluster assembly, as well as between NDUFA12 and its paralog, the assembly factor NDUFAF2.
Conclusions: The physical distance between subunits of complex I is a major correlate of the rate of protein evolution in the complex I matrix arm and is sufficient to infer parts of the complex's structure with high accuracy. The resulting evolutionary model predicts the positions of a number of subunits and assembly factors.
Figures

References
-
- Friedrich T, Abelmann A, Brors B, Guénebaut V, Kintscher L, Leonard K, Rasmussen T, Scheide D, Schlitt A, Schulte U. et al.Redox components and structure of the respiratory NADH:ubiquinone oxidoreductase (complex I) Biochim Biophys Acta. 1998;1365(1–2):215–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

