Effect of MHC and non-MHC donor/recipient genetic disparity on the outcome of allogeneic HCT
- PMID: 22859606
- PMCID: PMC3466963
- DOI: 10.1182/blood-2012-04-347286
Effect of MHC and non-MHC donor/recipient genetic disparity on the outcome of allogeneic HCT
Abstract
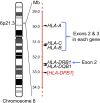
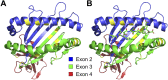
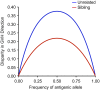
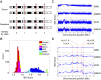
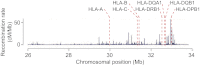
The outcome of allogeneic hematopoietic cell transplantation is influenced by donor/recipient genetic disparity at loci both inside and outside the MHC on chromosome 6p. Although disparity at loci within the MHC is the most important risk factor for the development of severe GVHD, disparity at loci outside the MHC that encode minor histocompatibility (H) antigens can elicit GVHD and GVL activity in donor/recipient pairs who are otherwise genetically identical across the MHC. Minor H antigens are created by sequence and structural variations within the genome. The enormous variation that characterizes the human genome suggests that the total number of minor H loci is probably large and ensures that all donor/recipient pairs, despite selection for identity at the MHC, will be mismatched for many minor H antigens. In addition to mismatch at minor H loci, unrelated donor/recipient pairs exhibit genetic disparity at numerous loci within the MHC, particularly HLA-DP, despite selection for identity at HLA-A, -B, -C, and -DRB1. Disparity at HLA-DP exists in 80% of unrelated pairs and clearly influences the outcome of unrelated hematopoietic cell transplantation; the magnitude of this effect probably exceeds that associated with disparity at any locus outside the MHC.
Figures


References
-
- Thomas E, Storb R, Clift RA, et al. Bone-marrow transplantation (first of two parts). N Engl J Med. 1975;292(16):832–843. - PubMed
-
- Thomas ED, Storb R, Clift RA, et al. Bone-marrow transplantation (second of two parts). N Engl J Med. 1975;292(17):895–902. - PubMed
-
- Bray RA, Hurley CK, Kamani NR, et al. National marrow donor program HLA matching guidelines for unrelated adult donor hematopoietic cell transplants. Biol Blood Marrow Transplant. 2008;14(9 Suppl):45–53. - PubMed
-
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–512. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

