Large-scale evaluation of automated clinical note de-identification and its impact on information extraction
- PMID: 22859645
- PMCID: PMC3555323
- DOI: 10.1136/amiajnl-2012-001012
Large-scale evaluation of automated clinical note de-identification and its impact on information extraction
Abstract
Objective: (1) To evaluate a state-of-the-art natural language processing (NLP)-based approach to automatically de-identify a large set of diverse clinical notes. (2) To measure the impact of de-identification on the performance of information extraction algorithms on the de-identified documents.
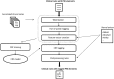
Material and methods: A cross-sectional study that included 3503 stratified, randomly selected clinical notes (over 22 note types) from five million documents produced at one of the largest US pediatric hospitals. Sensitivity, precision, F value of two automated de-identification systems for removing all 18 HIPAA-defined protected health information elements were computed. Performance was assessed against a manually generated 'gold standard'. Statistical significance was tested. The automated de-identification performance was also compared with that of two humans on a 10% subsample of the gold standard. The effect of de-identification on the performance of subsequent medication extraction was measured.
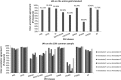
Results: The gold standard included 30 815 protected health information elements and more than one million tokens. The most accurate NLP method had 91.92% sensitivity (R) and 95.08% precision (P) overall. The performance of the system was indistinguishable from that of human annotators (annotators' performance was 92.15%(R)/93.95%(P) and 94.55%(R)/88.45%(P) overall while the best system obtained 92.91%(R)/95.73%(P) on same text). The impact of automated de-identification was minimal on the utility of the narrative notes for subsequent information extraction as measured by the sensitivity and precision of medication name extraction.
Discussion and conclusion: NLP-based de-identification shows excellent performance that rivals the performance of human annotators. Furthermore, unlike manual de-identification, the automated approach scales up to millions of documents quickly and inexpensively.
Conflict of interest statement
Figures


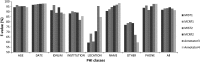

References
-
- Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 - PubMed
-
- Hicks J. The Potential of Claims Data to Support the Measurement of Health Care Quality. Santa Monica, CA: RAND Corporation, 2003
-
- Jha AK. The promise of electronic records: around the corner or down the road? JAMA 2011;306:880–1 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

