High resolution crystal structure of the endo-N-Acetyl-β-D-glucosaminidase responsible for the deglycosylation of Hypocrea jecorina cellulases
- PMID: 22859955
- PMCID: PMC3408457
- DOI: 10.1371/journal.pone.0040854
High resolution crystal structure of the endo-N-Acetyl-β-D-glucosaminidase responsible for the deglycosylation of Hypocrea jecorina cellulases
Abstract
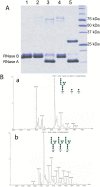
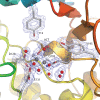
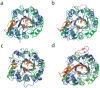
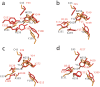
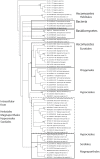
Endo-N-acetyl-β-D-glucosaminidases (ENGases) hydrolyze the glycosidic linkage between the two N-acetylglucosamine units that make up the chitobiose core of N-glycans. The endo-N-acetyl-β-D-glucosaminidases classified into glycoside hydrolase family 18 are small, bacterial proteins with different substrate specificities. Recently two eukaryotic family 18 deglycosylating enzymes have been identified. Here, the expression, purification and the 1.3Å resolution structure of the ENGase (Endo T) from the mesophilic fungus Hypocrea jecorina (anamorph Trichoderma reesei) are reported. Although the mature protein is C-terminally processed with removal of a 46 amino acid peptide, the protein has a complete (β/α)8 TIM-barrel topology. In the active site, the proton donor (E131) and the residue stabilizing the transition state (D129) in the substrate assisted catalysis mechanism are found in almost identical positions as in the bacterial GH18 ENGases: Endo H, Endo F1, Endo F3, and Endo BT. However, the loops defining the substrate-binding cleft vary greatly from the previously known ENGase structures, and the structures also differ in some of the α-helices forming the barrel. This could reflect the variation in substrate specificity between the five enzymes. This is the first three-dimensional structure of a eukaryotic endo-N-acetyl-β-D-glucosaminidase from glycoside hydrolase family 18. A glycosylation analysis of the cellulases secreted by a Hypocrea jecorina Endo T knock-out strain shows the in vivo function of the protein. A homology search and phylogenetic analysis show that the two known enzymes and their homologues form a large but separate cluster in subgroup B of the fungal chitinases. Therefore the future use of a uniform nomenclature is proposed.
Conflict of interest statement
Figures
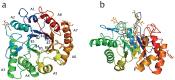

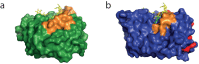
References
-
- Tarentino AL, Plummer TH Jr, Maley F (1974) The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. J Biol Chem. 249: 818–824. - PubMed
-
- Trimble RB, Tarentino AL (1991) Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J Biol Chem 266: 1646–1651. - PubMed
-
- Collin M, Fischetti VA (2004) A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J Biol Chem. 279: 22558–22570. - PubMed
-
- Genomics JCFS (2010) Crystal structure of an endo-beta-N-acetylglucosaminidase BT_3987 from Bacteroides thetaiotaomicron at 1.55 Å resolution. (PDB ID 3POH)..
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

