Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene
- PMID: 22859989
- PMCID: PMC3409227
- DOI: 10.1371/journal.pone.0041484
Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene
Abstract
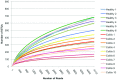
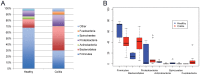
The intestinal tract houses one of the richest and most complex microbial populations on the planet, and plays a critical role in health and a wide range of diseases. Limited studies using new sequencing technologies in horses are available. The objective of this study was to characterize the fecal microbiome of healthy horses and to compare the fecal microbiome of healthy horses to that of horses with undifferentiated colitis. A total of 195,748 sequences obtained from 6 healthy horses and 10 horses affected by undifferentiated colitis were analyzed. Firmicutes predominated (68%) among healthy horses followed by Bacteroidetes (14%) and Proteobacteria (10%). In contrast, Bacteroidetes (40%) was the most abundant phylum among horses with colitis, followed by Firmicutes (30%) and Proteobacteria (18%). Healthy horses had a significantly higher relative abundance of Actinobacteria and Spirochaetes while horses with colitis had significantly more Fusobacteria. Members of the Clostridia class were more abundant in healthy horses. Members of the Lachnospiraceae family were the most frequently shared among healthy individuals. The species richness reported here indicates the complexity of the equine intestinal microbiome. The predominance of Clostridia demonstrates the importance of this group of bacteria in healthy horses. The marked differences in the microbiome between healthy horses and horses with colitis indicate that colitis may be a disease of gut dysbiosis, rather than one that occurs simply through overgrowth of an individual pathogen.
Conflict of interest statement
Figures


References
-
- Argenzio RA, Southworth M, Stevens CE (1974) Sites of organic acid production and absorption in the equine gastrointestinal tract. Am J Physiol 226: 1043–1050. - PubMed
-
- Glinsky MJ, Smith RM, Spires HR, Davis CL (1976) Measurement of Volatile Fatty-Acid Production-Rates in Cecum of Pony. Anim Sci 42: 1465–1470. - PubMed
-
- Al Jassim RA, Andrews FM (2009) The bacterial community of the horse gastrointestinal tract and its relation to fermentative acidosis, laminitis, colic, and stomach ulcers. Vet Clin North Am Equine Pract 25: 199–215. - PubMed
-
- Chapman AM (2001) Acute diarrhea in hospitalized horses. Vet Clin North Am Equine Pract 25: 363–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

