Identification of pluripotent and adult stem cell genes unrelated to cell cycle and associated with poor prognosis in multiple myeloma
- PMID: 22860071
- PMCID: PMC3409163
- DOI: 10.1371/journal.pone.0042161
Identification of pluripotent and adult stem cell genes unrelated to cell cycle and associated with poor prognosis in multiple myeloma
Abstract
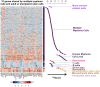
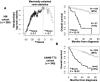
Gene expression-based scores used to predict risk in cancer frequently include genes coding for DNA replication, repair or recombination. Using two independent cohorts of 206 and 345 previously-untreated patients with Multiple Myeloma (MM), we identified 50 cell cycle-unrelated genes overexpressed in multiple myeloma cells (MMCs) compared to normal human proliferating plasmablasts and non-proliferating bone marrow plasma cells and which have prognostic value for overall survival. Thirty-seven of these 50 myeloma genes (74%) were enriched in genes overexpressed in one of 3 normal human stem cell populations--pluripotent (18), hematopoietic (10) or mesenchymal stem cells (9)--and only three genes were enriched in one of 5 populations of differentiated cells (memory B lymphocytes, T lymphocytes, polymorphonuclear cells, monocytes, osteoclasts). These 37 genes shared by MMCs and adult or pluripotent stem cells were used to build a stem cell score ((SC)score), which proved to be strongly prognostic in the 2 independent cohorts of patients compared to other gene expression-based risk scores or usual clinical scores using multivariate Cox analysis. This finding highlights cell cycle-unrelated prognostic genes shared by myeloma cells and normal stem cells, whose products might be important for normal and malignant stem cell biology.
Conflict of interest statement
Figures

References
-
- Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, et al. (2010) A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 116: e56–65. - PubMed
-
- Shaughnessy JD Jr, Zhan F, Burington BE, Huang Y, Colla S, et al. (2007) A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109: 2276–2284. - PubMed
-
- Decaux O, Lode L, Magrangeas F, Charbonnel C, Gouraud W, et al. (2008) Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myelome. J Clin Oncol 26: 4798–4805. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

