Mapping paratope on antithrombotic antibody 6B4 to epitope on platelet glycoprotein Ibalpha via molecular dynamic simulations
- PMID: 22860101
- PMCID: PMC3408434
- DOI: 10.1371/journal.pone.0042263
Mapping paratope on antithrombotic antibody 6B4 to epitope on platelet glycoprotein Ibalpha via molecular dynamic simulations
Abstract
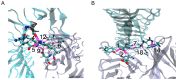
Binding of platelet receptor glycoprotein Ibα (GPIbα) to the A1 domain of von Willebrand factor (vWF) is a critical step in both physiologic hemostasis and pathologic thrombosis, for initiating platelet adhesion to subendothelium of blood vessels at sites of vascular injury. Gain-of-function mutations in GPIbα contribute to an abnormally high-affinity binding of platelets to vWF and can lead to thrombosis, an accurate complication causing heart attack and stroke. Of various antithrombotic monoclonal antibodies (mAbs) targeting human GPIbα, 6B4 is a potent one to inhibit the interaction between GPIbα and vWF-A1 under static and flow conditions. Mapping paratope to epitope with mutagenesis experiments, a traditional route in researches of these antithrombotic mAbs, is usually expensive and time-consuming. Here, we suggested a novel computational procedure, which combines with homology modeling, rigid body docking, free and steered molecular dynamics (MD) simulations, to identify key paratope residues on 6B4 and their partners on GPIbα, with hypothesis that the stable hydrogen bonds and salt bridges are the important linkers between paratope and epitope residues. Based on a best constructed model of 6B4 bound with GPIbα, the survival ratios and rupture times of all detected hydrogen bonds and salt bridges in binding site were examined via free and steered MD simulations and regarded as indices of thermal and mechanical stabilizations of the bonds, respectively. Five principal paratope residues with their partners were predicted with their high survival ratios and/or long rupture times of involved hydrogen bonds, or with their hydrogen bond stabilization indices ranked in top 5. Exciting, the present results were in good agreement with previous mutagenesis experiment data, meaning a wide application prospect of our novel computational procedure on researches of molecular of basis of ligand-receptor interactions, various antithrombotic mAbs and other antibodies as well as theoretically design of biomolecular drugs.
Conflict of interest statement
Figures

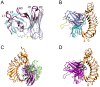



References
-
- Savage B, Saldivar E, Ruggeri ZM (1996) Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell 84: 289–297. - PubMed
-
- Ruggeri ZM (2002) Platelets in atherothrombosis. Nat Med 8: 1227–1234. - PubMed
-
- Russell SD, Roth GJ (1993) Pseudo-von Willebrand disease: a mutation in the platelet glycoprotein Ib alpha gene associated with a hyperactive surface receptor. Blood 81: 1787–1791. - PubMed
-
- Sadler JE (2005) New concepts in von Willebrand disease. Annu Rev Med 56: 173–191. - PubMed
-
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, et al. (2002) Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science 297: 1176–1179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

