A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product
- PMID: 22860154
- PMCID: PMC3409129
- DOI: 10.1371/journal.pntd.0001777
A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product
Abstract
Background: Trypanosoma cruzi is the causative agent of Chagas' Disease. The parasite has a complex population structure, with six major evolutionary lineages, some of which have apparently resulted from ancestral hybridization events. Because there are important biological differences between these lineages, strain typing methods are essential to study the T. cruzi species. Currently, there are a number of typing methods available for T. cruzi, each with its own advantages and disadvantages. However, most of these methods are based on the amplification of a variable number of loci.
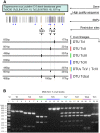
Methodology/principal findings: We present a simple typing assay for T. cruzi, based on the amplification of a single polymorphic locus: the TcSC5D gene. When analyzing sequences from this gene (a putative lathosterol/episterol oxidase) we observed a number of interesting polymorphic sites, including 1 tetra-allelic, and a number of informative tri- and bi-allelic SNPs. Furthermore, some of these SNPs were located within the recognition sequences of two commercially available restriction enzymes. A double digestion with these enzymes generates a unique restriction pattern that allows a simple classification of strains in six major groups, corresponding to DTUs TcI-TcIV, the recently proposed Tcbat lineage, and TcV/TcVI (as a group). Direct sequencing of the amplicon allows the classification of strains into seven groups, including the six currently recognized evolutionary lineages, by analyzing only a few discriminant polymorphic sites.
Conclusions/significance: Based on these findings we propose a simple typing assay for T. cruzi that requires a single PCR amplification followed either by restriction fragment length polymorphism analysis, or direct sequencing. In the panel of strains tested, the sequencing-based method displays equivalent inter-lineage resolution to recent multi- locus sequence typing assays. Due to their simplicity and low cost, the proposed assays represent a good alternative to rapidly screen strain collections, providing the cornerstone for the development of robust typing strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rassi A, Rassi A, Marin Neto JA (2010) Chagas disease. Lancet 375: 1388–1402. - PubMed
-
- Dvorak JA (1984) The natural heterogeneity of Trypanosoma cruzi: biological and medical implications. J Cell Biochem 24: 357–371. - PubMed
-
- Tibayrenc M, Ayala FJ (1998) Isozyme variability in Trypanosoma cruzi, the agent of Chagas' disease: Genetical, taxomonical, and epidemiological significance. Evolution 42: 277–292. - PubMed
-
- Tibayrenc M, Ayala FJ (2002) The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol 18: 405–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

