p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance
- PMID: 22861165
- PMCID: PMC3526895
- DOI: 10.1089/ars.2012.4615
p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance
Abstract
Aims: The transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1 α (PPARGC1A or PGC-1α) is a powerful controller of cell metabolism and assures the balance between the production and the scavenging of pro-oxidant molecules by coordinating mitochondrial biogenesis and the expression of antioxidants. However, even though a huge amount of data referring to the role of PGC-1α is available, the molecular mechanisms of its regulation at the transcriptional level are not completely understood. In the present report, we aim at characterizing whether the decrease of antioxidant glutathione (GSH) modulates PGC-1α expression and its downstream metabolic pathways.
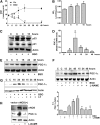
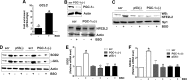
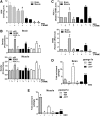
Results: We found that upon GSH shortage, induced either by its chemical depletion or by metabolic stress (i.e., fasting), p53 binds to the PPARGC1A promoter of both human and mouse genes, and this event is positively related to increased PGC-1α expression. This effect was abrogated by inhibiting nitric oxide (NO) synthase or guanylate cyclase, implicating NO/cGMP signaling in such a process. We show that p53-mediated PGC-1α upregulation is directed to potentiate the antioxidant defense through nuclear factor (erythroid-derived 2)-like2 (NFE2L2)-mediated expression of manganese superoxide dismutase (SOD2) and γ-glutamylcysteine ligase without modulating mitochondrial biogenesis.
Innovation and conclusions: We outlined a new NO-dependent signaling axis responsible for survival antioxidant response upon mild metabolic stress (fasting) and/or oxidative imbalance (GSH depletion). Such signaling axis could become the cornerstone for new pharmacological or dietary approaches for improving antioxidant response during ageing and human pathologies associated with oxidative stress.
Figures






References
-
- Aquilano K. Baldelli S. Cardaci S. Rotilio G. Ciriolo MR. Nitric oxide is the primary mediator of cytotoxicity induced by GSH depletion in neuronal cells. J Cell Sci. 2011;124:1043–1054. - PubMed
-
- Aquilano K. Vigilanza P. Baldelli S. Pagliei B. Rotilio G. Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. - PMC - PubMed
-
- Aquilano K. Vigilanza P. Rotilio G. Ciriolo MR. Mitochondrial damage due to SOD1 deficiency in SH-SY5Y neuroblastoma cells: a rationale for the redundancy of SOD1. FASEB J. 2006;20:1683–1685. - PubMed
-
- Baldelli S. Aquilano K. Rotilio G. Ciriolo MR. Glutathione and copper, zinc superoxide dismutase are modulated by overexpression of neuronal nitric oxide synthase. Int J Biochem Cell Biol. 2008;40:2660–2670. - PubMed
-
- Baldelli S. Aquilano K. Rotilio G. Ciriolo MR. Neuronal nitric oxide synthase interacts with Sp1 through the PDZ domain inhibiting Sp1-mediated copper-zinc superoxide dismutase expression. Int J Biochem Cell Biol. 2011;43:163–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

