Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development
- PMID: 22863006
- PMCID: PMC3700733
- DOI: 10.1016/j.cell.2012.05.048
Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development
Abstract
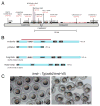
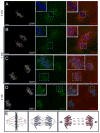
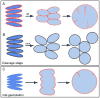
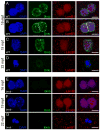
To accommodate the large cells following zygote formation, early blastomeres employ modified cell divisions. Karyomeres are one such modification, mitotic intermediates wherein individual chromatin masses are surrounded by nuclear envelope; the karyomeres then fuse to form a single mononucleus. We identified brambleberry, a maternal-effect zebrafish mutant that disrupts karyomere fusion, resulting in formation of multiple micronuclei. As karyomeres form, Brambleberry protein localizes to the nuclear envelope, with prominent puncta evident near karyomere-karyomere interfaces corresponding to membrane fusion sites. brambleberry corresponds to an unannotated gene with similarity to Kar5p, a protein that participates in nuclear fusion in yeast. We also demonstrate that Brambleberry is required for pronuclear fusion following fertilization in zebrafish. Our studies provide insight into the machinery required for karyomere fusion and suggest that specialized proteins are necessary for proper nuclear division in large dividing blastomeres.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
-
- Abramoff MD, Magalhaes PJ, JRS . Image Processing with ImageJ. Biophotonics International; 2004.
-
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. - PubMed
-
- Balakier H, Cadesky K. The frequency and developmental capability of human embryos containing multinucleated blastomeres. Hum Reprod. 1997;12:800–804. - PubMed
-
- Baur T, Ramadan K, Schlundt A, Kartenbeck J, Meyer HH. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J Cell Sci. 2007;120:2895–2903. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- F32 GM080926/GM/NIGMS NIH HHS/United States
- 5F32GM77835/GM/NIGMS NIH HHS/United States
- F32 GM077835/GM/NIGMS NIH HHS/United States
- T32 HD007516/HD/NICHD NIH HHS/United States
- R01 HD050901/HD/NICHD NIH HHS/United States
- R01 HD065600/HD/NICHD NIH HHS/United States
- T32-HD007516/HD/NICHD NIH HHS/United States
- R21 HD062952/HD/NICHD NIH HHS/United States
- R01HD050901/HD/NICHD NIH HHS/United States
- 1F32GM080926/GM/NIGMS NIH HHS/United States
- R01HD065600/HD/NICHD NIH HHS/United States
- R21HD062952/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

