Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium
- PMID: 22863011
- PMCID: PMC3624753
- DOI: 10.1016/j.cell.2012.06.026
Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium
Abstract
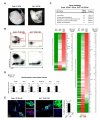
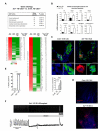
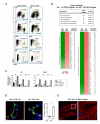
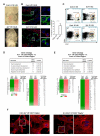
Endothelium in embryonic hematopoietic tissues generates hematopoietic stem/progenitor cells; however, it is unknown how its unique potential is specified. We show that transcription factor Scl/Tal1 is essential for both establishing the hematopoietic transcriptional program in hemogenic endothelium and preventing its misspecification to a cardiomyogenic fate. Scl(-/-) embryos activated a cardiac transcriptional program in yolk sac endothelium, leading to the emergence of CD31+Pdgfrα+ cardiogenic precursors that generated spontaneously beating cardiomyocytes. Ectopic cardiogenesis was also observed in Scl(-/-) hearts, where the disorganized endocardium precociously differentiated into cardiomyocytes. Induction of mosaic deletion of Scl in Scl(fl/fl)Rosa26Cre-ER(T2) embryos revealed a cell-intrinsic, temporal requirement for Scl to prevent cardiomyogenesis from endothelium. Scl(-/-) endothelium also upregulated the expression of Wnt antagonists, which promoted rapid cardiomyocyte differentiation of ectopic cardiogenic cells. These results reveal unexpected plasticity in embryonic endothelium such that loss of a single master regulator can induce ectopic cardiomyogenesis from endothelial cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
-
- Ahnfelt-Ronne J, Jorgensen MC, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An improved method for three-dimensional reconstruction of protein expression patterns in intact mouse and chicken embryos and organs. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2007;55:925–930. - PubMed
-
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:759–767. - PubMed
-
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

