A synthetic biology framework for programming eukaryotic transcription functions
- PMID: 22863014
- PMCID: PMC3653585
- DOI: 10.1016/j.cell.2012.05.045
A synthetic biology framework for programming eukaryotic transcription functions
Abstract
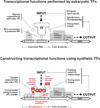
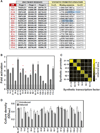
Eukaryotic transcription factors (TFs) perform complex and combinatorial functions within transcriptional networks. Here, we present a synthetic framework for systematically constructing eukaryotic transcription functions using artificial zinc fingers, modular DNA-binding domains found within many eukaryotic TFs. Utilizing this platform, we construct a library of orthogonal synthetic transcription factors (sTFs) and use these to wire synthetic transcriptional circuits in yeast. We engineer complex functions, such as tunable output strength and transcriptional cooperativity, by rationally adjusting a decomposed set of key component properties, e.g., DNA specificity, affinity, promoter design, protein-protein interactions. We show that subtle perturbations to these properties can transform an individual sTF between distinct roles (activator, cooperative factor, inhibitory factor) within a transcriptional complex, thus drastically altering the signal processing behavior of multi-input systems. This platform provides new genetic components for synthetic biology and enables bottom-up approaches to understanding the design principles of eukaryotic transcriptional complexes and networks.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. - PubMed
-
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. - PubMed
-
- Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

