Investigating metabotropic glutamate receptor 5 allosteric modulator cooperativity, affinity, and agonism: enriching structure-function studies and structure-activity relationships
- PMID: 22863693
- PMCID: PMC3477233
- DOI: 10.1124/mol.112.080531
Investigating metabotropic glutamate receptor 5 allosteric modulator cooperativity, affinity, and agonism: enriching structure-function studies and structure-activity relationships
Abstract
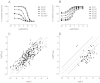
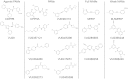
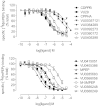
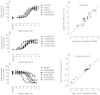
Drug discovery programs increasingly are focusing on allosteric modulators as a means to modify the activity of G protein-coupled receptor (GPCR) targets. Allosteric binding sites are topographically distinct from the endogenous ligand (orthosteric) binding site, which allows for co-occupation of a single receptor with the endogenous ligand and an allosteric modulator that can alter receptor pharmacological characteristics. Negative allosteric modulators (NAMs) inhibit and positive allosteric modulators (PAMs) enhance the affinity and/or efficacy of orthosteric agonists. Established approaches for estimation of affinity and efficacy values for orthosteric ligands are not appropriate for allosteric modulators, and this presents challenges for fully understanding the actions of novel modulators of GPCRs. Metabotropic glutamate receptor 5 (mGlu(5)) is a family C GPCR for which a large array of allosteric modulators have been identified. We took advantage of the many tools for probing allosteric sites on mGlu(5) to validate an operational model of allosterism that allows quantitative estimation of modulator affinity and cooperativity values. Affinity estimates derived from functional assays fit well with affinities measured in radioligand binding experiments for both PAMs and NAMs with diverse chemical scaffolds and varying degrees of cooperativity. We observed modulation bias for PAMs when we compared mGlu(5)-mediated Ca(2+) mobilization and extracellular signal-regulated kinase 1/2 phosphorylation data. Furthermore, we used this model to quantify the effects of mutations that reduce binding or potentiation by PAMs. This model can be applied to PAM and NAM potency curves in combination with maximal fold-shift data to derive reliable estimates of modulator affinities.
Figures





References
-
- Chen Y, Goudet C, Pin JP, Conn PJ. (2008) N-{4-Chloro-2-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-hydroxybenzamide (CPPHA) acts through a novel site as a positive allosteric modulator of group 1 metabotropic glutamate receptors. Mol Pharmacol 73:909–918 - PubMed
-
- Chen Y, Nong Y, Goudet C, Hemstapat K, de Paulis T, Pin JP, Conn PJ. (2007) Interaction of novel positive allosteric modulators of metabotropic glutamate receptor 5 with the negative allosteric antagonist site is required for potentiation of receptor responses. Mol Pharmacol 71:1389–1398 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32-NS071746/NS/NINDS NIH HHS/United States
- R01 MH062646/MH/NIMH NIH HHS/United States
- U54 MH084659/MH/NIMH NIH HHS/United States
- R01 NS031373/NS/NINDS NIH HHS/United States
- 1R01-DA023947/DA/NIDA NIH HHS/United States
- 5U54-MH84659-03/MH/NIMH NIH HHS/United States
- F32 MH088234/MH/NIMH NIH HHS/United States
- F32-MH088234-02/MH/NIMH NIH HHS/United States
- U19 MH097056/MH/NIMH NIH HHS/United States
- 5U54-MH84659-03S1/MH/NIMH NIH HHS/United States
- 2R01-NS031373-16A2/NS/NINDS NIH HHS/United States
- F32 NS071746/NS/NINDS NIH HHS/United States
- 2R01-MH062646-13/MH/NIMH NIH HHS/United States
- R01 DA023947/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

