Human CST promotes telomere duplex replication and general replication restart after fork stalling
- PMID: 22863775
- PMCID: PMC3433780
- DOI: 10.1038/emboj.2012.215
Human CST promotes telomere duplex replication and general replication restart after fork stalling
Abstract
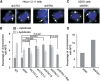
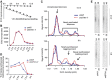
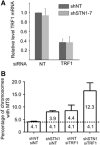
Mammalian CST (CTC1-STN1-TEN1) associates with telomeres and depletion of CTC1 or STN1 causes telomere defects. However, the function of mammalian CST remains poorly understood. We show here that depletion of CST subunits leads to both telomeric and non-telomeric phenotypes associated with DNA replication defects. Stable knockdown of CTC1 or STN1 increases the incidence of anaphase bridges and multi-telomeric signals, indicating genomic and telomeric instability. STN1 knockdown also delays replication through the telomere indicating a role in replication fork passage through this natural barrier. Furthermore, we find that STN1 plays a novel role in genome-wide replication restart after hydroxyurea (HU)-induced replication fork stalling. STN1 depletion leads to reduced EdU incorporation after HU release. However, most forks rapidly resume replication, indicating replisome integrity is largely intact and STN1 depletion has little effect on fork restart. Instead, STN1 depletion leads to a decrease in new origin firing. Our findings suggest that CST rescues stalled replication forks during conditions of replication stress, such as those found at natural replication barriers, likely by facilitating dormant origin firing.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
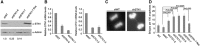


Similar articles
-
Human CST abundance determines recovery from diverse forms of DNA damage and replication stress.Cell Cycle. 2014;13(22):3488-98. doi: 10.4161/15384101.2014.964100. Cell Cycle. 2014. PMID: 25483097 Free PMC article.
-
Human TEN1 maintains telomere integrity and functions in genome-wide replication restart.J Biol Chem. 2013 Oct 18;288(42):30139-30150. doi: 10.1074/jbc.M113.493478. Epub 2013 Sep 11. J Biol Chem. 2013. PMID: 24025336 Free PMC article.
-
Human CST has independent functions during telomere duplex replication and C-strand fill-in.Cell Rep. 2012 Nov 29;2(5):1096-103. doi: 10.1016/j.celrep.2012.10.007. Epub 2012 Nov 8. Cell Rep. 2012. PMID: 23142664 Free PMC article.
-
Emerging roles of CST in maintaining genome stability and human disease.Front Biosci (Landmark Ed). 2018 Mar 1;23(8):1564-1586. doi: 10.2741/4661. Front Biosci (Landmark Ed). 2018. PMID: 29293451 Free PMC article. Review.
-
CST in maintaining genome stability: Beyond telomeres.DNA Repair (Amst). 2021 Jun;102:103104. doi: 10.1016/j.dnarep.2021.103104. Epub 2021 Mar 22. DNA Repair (Amst). 2021. PMID: 33780718 Free PMC article. Review.
Cited by
-
Suppression of STN1 enhances the cytotoxicity of chemotherapeutic agents in cancer cells by elevating DNA damage.Oncol Lett. 2016 Aug;12(2):800-808. doi: 10.3892/ol.2016.4676. Epub 2016 Jun 2. Oncol Lett. 2016. PMID: 27446354 Free PMC article.
-
Developmentally Programmed Switches in DNA Replication: Gene Amplification and Genome-Wide Endoreplication in Tetrahymena.Microorganisms. 2023 Feb 16;11(2):491. doi: 10.3390/microorganisms11020491. Microorganisms. 2023. PMID: 36838456 Free PMC article.
-
Consequences of telomere replication failure: the other end-replication problem.Trends Biochem Sci. 2022 Jun;47(6):506-517. doi: 10.1016/j.tibs.2022.03.013. Epub 2022 Apr 16. Trends Biochem Sci. 2022. PMID: 35440402 Free PMC article. Review.
-
Structural Analysis and Conformational Dynamics of STN1 Gene Mutations Involved in Coat Plus Syndrome.Front Mol Biosci. 2019 Jun 12;6:41. doi: 10.3389/fmolb.2019.00041. eCollection 2019. Front Mol Biosci. 2019. PMID: 31245382 Free PMC article.
-
The structure of human CST reveals a decameric assembly bound to telomeric DNA.Science. 2020 Jun 5;368(6495):1081-1085. doi: 10.1126/science.aaz9649. Science. 2020. PMID: 32499435 Free PMC article.
References
-
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O'Sullivan J, Leibundgut EO, Muter J, Abdel-Salem GM, Babul-Hirji R, Baxter P, Berger A, Bonafe L, Brunstom-Hernandez JE, Buckard JA, Chitayat D et al. (2012) Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44: 338–342 - PubMed
-
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

