Going viral: next-generation sequencing applied to phage populations in the human gut
- PMID: 22864264
- PMCID: PMC3596094
- DOI: 10.1038/nrmicro2853
Going viral: next-generation sequencing applied to phage populations in the human gut
Abstract
Over the past decade, researchers have begun to characterize viral diversity using metagenomic methods. These studies have shown that viruses, the majority of which infect bacteria, are probably the most genetically diverse components of the biosphere. Here, we briefly review the incipient rise of a phage biology renaissance, which has been catalysed by advances in next-generation sequencing. We explore how work characterizing phage diversity and lifestyles in the human gut is changing our view of ourselves as supra-organisms. Finally, we discuss how a renewed appreciation of phage dynamics may yield new applications for phage therapies designed to manipulate the structure and functions of our gut microbiomes.
Figures
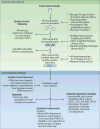

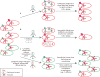
References
-
- Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. - PubMed
-
- Carins J, Stent GS, Watson JD. Phage and the Origins of Molecular Biology. Cold Spring Harbor Laboratory Press; Plainview, NY: 1992.
-
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

