Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.)
- PMID: 22865733
- PMCID: PMC3454892
- DOI: 10.1534/genetics.112.141473
Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.)
Abstract
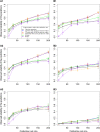
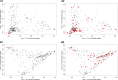
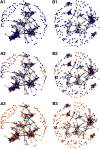
Genomic selection refers to the use of genotypic information for predicting breeding values of selection candidates. A prediction formula is calibrated with the genotypes and phenotypes of reference individuals constituting the calibration set. The size and the composition of this set are essential parameters affecting the prediction reliabilities. The objective of this study was to maximize reliabilities by optimizing the calibration set. Different criteria based on the diversity or on the prediction error variance (PEV) derived from the realized additive relationship matrix-best linear unbiased predictions model (RA-BLUP) were used to select the reference individuals. For the latter, we considered the mean of the PEV of the contrasts between each selection candidate and the mean of the population (PEVmean) and the mean of the expected reliabilities of the same contrasts (CDmean). These criteria were tested with phenotypic data collected on two diversity panels of maize (Zea mays L.) genotyped with a 50k SNPs array. In the two panels, samples chosen based on CDmean gave higher reliabilities than random samples for various calibration set sizes. CDmean also appeared superior to PEVmean, which can be explained by the fact that it takes into account the reduction of variance due to the relatedness between individuals. Selected samples were close to optimality for a wide range of trait heritabilities, which suggests that the strategy presented here can efficiently sample subsets in panels of inbred lines. A script to optimize reference samples based on CDmean is available on request.
Figures


References
-
- Albrecht T., Wimmer V., Auinger H.-J., Erbe M., Knaak C., et al. , 2011. Genome-based prediction of testcross values in maize. Theor. Appl. Genet. 123: 339–350 - PubMed
-
- Astle W., Balding D. J., 2009. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24: 451–471
-
- Atkinson A. C., Donev A. N., Tobias R. D., 2007. Optimum Experimental Designs, With SAS. Clarendon Press, Oxford
-
- Bernardo R., Yu J., 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47: 1082
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

