mRNA localization and translational control in Drosophila oogenesis
- PMID: 22865893
- PMCID: PMC3475173
- DOI: 10.1101/cshperspect.a012294
mRNA localization and translational control in Drosophila oogenesis
Abstract
Localization of an mRNA species to a particular subcellular region can complement translational control mechanisms to produce a restricted spatial distribution of the protein it encodes. mRNA localization has been studied most in asymmetric cells such as budding yeast, early embryos, and neurons, but the process is likely to be more widespread. This article reviews the current state of knowledge about the mechanisms of mRNA localization and its functions in early embryonic development, focusing on Drosophila where the relevant knowledge is most advanced. Links between mRNA localization and translational control mechanisms also are examined.
Figures
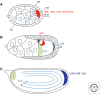
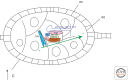
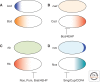
References
-
- Arnott S, Hukins DW, Dover SD 1972. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun 48: 1392–1399 - PubMed
-
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD 1999. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J 18: 2610–2620 - PMC - PubMed
-
- Bastock R, St Johnston D 2008. Drosophila oogenesis. Curr Biol 18: R1082–R1087 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
