Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics
- PMID: 22865924
- PMCID: PMC3494192
- DOI: 10.1074/mcp.O112.020131
Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics
Abstract
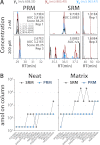
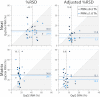
Selected reaction monitoring on a triple quadrupole mass spectrometer is currently experiencing a renaissance within the proteomics community for its, as yet, unparalleled ability to characterize and quantify a set of proteins reproducibly, completely, and with high sensitivity. Given the immense benefit that high resolution and accurate mass instruments have brought to the discovery proteomics field, we wondered if highly accurate mass measurement capabilities could be leveraged to provide benefits in the targeted proteomics domain as well. Here, we propose a new targeted proteomics paradigm centered on the use of next generation, quadrupole-equipped high resolution and accurate mass instruments: parallel reaction monitoring (PRM). In PRM, the third quadrupole of a triple quadrupole is substituted with a high resolution and accurate mass mass analyzer to permit the parallel detection of all target product ions in one, concerted high resolution mass analysis. We detail the analytical performance of the PRM method, using a quadrupole-equipped bench-top Orbitrap MS, and draw a performance comparison to selected reaction monitoring in terms of run-to-run reproducibility, dynamic range, and measurement accuracy. In addition to requiring minimal upfront method development and facilitating automated data analysis, PRM yielded quantitative data over a wider dynamic range than selected reaction monitoring in the presence of a yeast background matrix because of PRM's high selectivity in the mass-to-charge domain. With achievable linearity over the quantifiable dynamic range found to be statistically equal between the two methods, our investigation suggests that PRM will be a promising new addition to the quantitative proteomics toolbox.
Figures


Comment in
-
Targeting with PRM.Nat Methods. 2012 Oct;9(10):950. doi: 10.1038/nmeth.2193. Nat Methods. 2012. PMID: 23193583 No abstract available.
References
-
- Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 - PubMed
-
- Wu C. C., MacCoss M. J., Howell K. E., Matthews D. E., Yates J. R. (2004) Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76, 4951–4959 - PubMed
-
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

