Learning score function parameters for improved spectrum identification in tandem mass spectrometry experiments
- PMID: 22866926
- PMCID: PMC3436966
- DOI: 10.1021/pr300234m
Learning score function parameters for improved spectrum identification in tandem mass spectrometry experiments
Abstract
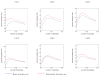
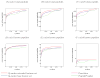
The identification of proteins from spectra derived from a tandem mass spectrometry experiment involves several challenges: matching each observed spectrum to a peptide sequence, ranking the resulting collection of peptide-spectrum matches, assigning statistical confidence estimates to the matches, and identifying the proteins. The present work addresses algorithms to rank peptide-spectrum matches. Many of these algorithms, such as PeptideProphet, IDPicker, or Q-ranker, follow a similar methodology that includes representing peptide-spectrum matches as feature vectors and using optimization techniques to rank them. We propose a richer and more flexible feature set representation that is based on the parametrization of the SEQUEST XCorr score and that can be used by all of these algorithms. This extended feature set allows a more effective ranking of the peptide-spectrum matches based on the target-decoy strategy, in comparison to a baseline feature set devoid of these XCorr-based features. Ranking using the extended feature set gives 10-40% improvement in the number of distinct peptide identifications relative to a range of q-value thresholds. While this work is inspired by the model of the theoretical spectrum and the similarity measure between spectra used specifically by SEQUEST, the method itself can be applied to the output of any database search. Further, our approach can be trivially extended beyond XCorr to any linear operator that can serve as similarity score between experimental spectra and peptide sequences.
Figures




References
-
- Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nature Methods. 2007;4(10):787–797. - PubMed
-
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. - PubMed
-
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identification made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. - PubMed
-
- Choi H, Nesvizhskii AI. Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. Journal of Proteome Research. 2008;7(1):254–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

