Epigenetic mechanisms in neurological disease
- PMID: 22869198
- PMCID: PMC3596876
- DOI: 10.1038/nm.2828
Epigenetic mechanisms in neurological disease
Abstract
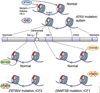
The exploration of brain epigenomes, which consist of various types of DNA methylation and covalent histone modifications, is providing new and unprecedented insights into the mechanisms of neural development, neurological disease and aging. Traditionally, chromatin defects in the brain were considered static lesions of early development that occurred in the context of rare genetic syndromes, but it is now clear that mutations and maladaptations of the epigenetic machinery cover a much wider continuum that includes adult-onset neurodegenerative disease. Here, we describe how recent advances in neuroepigenetics have contributed to an improved mechanistic understanding of developmental and degenerative brain disorders, and we discuss how they could influence the development of future therapies for these conditions.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

