Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas
- PMID: 22869205
- PMCID: PMC3443254
- DOI: 10.18632/oncotarget.588
Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas
Abstract
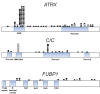
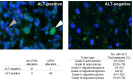
Mutations in the critical chromatin modifier ATRX and mutations in CIC and FUBP1, which are potent regulators of cell growth, have been discovered in specific subtypes of gliomas, the most common type of primary malignant brain tumors. However, the frequency of these mutations in many subtypes of gliomas, and their association with clinical features of the patients, is poorly understood. Here we analyzed these loci in 363 brain tumors. ATRX is frequently mutated in grade II-III astrocytomas (71%), oligoastrocytomas (68%), and secondary glioblastomas (57%), and ATRX mutations are associated with IDH1 mutations and with an alternative lengthening of telomeres phenotype. CIC and FUBP1 mutations occurred frequently in oligodendrogliomas (46% and 24%, respectively) but rarely in astrocytomas or oligoastrocytomas ( more than 10%). This analysis allowed us to define two highly recurrent genetic signatures in gliomas: IDH1/ATRX (I-A) and IDH1/CIC/FUBP1 (I-CF). Patients with I-CF gliomas had a significantly longer median overall survival (96 months) than patients with I-A gliomas (51 months) and patients with gliomas that did not harbor either signature (13 months). The genetic signatures distinguished clinically distinct groups of oligoastrocytoma patients, which usually present a diagnostic challenge, and were associated with differences in clinical outcome even among individual tumor types. In addition to providing new clues about the genetic alterations underlying gliomas, the results have immediate clinical implications, providing a tripartite genetic signature that can serve as a useful adjunct to conventional glioma classification that may aid in prognosis, treatment selection, and therapeutic trial design.
Figures



References
-
- Wen PY, Kesari S. Malignant glioma in adults (vol 359, pg 492, 2008) New Engl J Med. 2008;359(8):877–877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 CA043460/CA/NCI NIH HHS/United States
- P50 NS020023/NS/NINDS NIH HHS/United States
- 5P50 NS20023/NS/NINDS NIH HHS/United States
- R01 CA140316/CA/NCI NIH HHS/United States
- R37 CA057345/CA/NCI NIH HHS/United States
- R01 CA129825/CA/NCI NIH HHS/United States
- CA129825/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 CA 011898/CA/NCI NIH HHS/United States
- 5R01-CA140316/CA/NCI NIH HHS/United States
- 5P50 CA108785/CA/NCI NIH HHS/United States
- CA057345,/CA/NCI NIH HHS/United States
- R01 CA057345/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

