Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes
- PMID: 22869554
- PMCID: PMC3494202
- DOI: 10.1074/mcp.M112.019331
Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes
Abstract
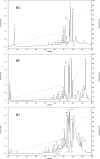
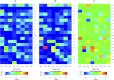
Snake venom proteomes/peptidomes are highly complex and maintenance of their integrity within the gland lumen is crucial for the expression of toxin activities. There has been considerable progress in the field of venom proteomics, however, peptidomics does not progress as fast, because of the lack of comprehensive venom sequence databases for analysis of MS data. Therefore, in many cases venom peptides have to be sequenced manually by MS/MS analysis or Edman degradation. This is critical for rare snake species, as is the case of Bothrops cotiara (BC) and B. fonsecai (BF), which are regarded as near threatened with extinction. In this study we conducted a comprehensive analysis of the venom peptidomes of BC, BF, and B. jararaca (BJ) using a combination of solid-phase extraction and reversed-phase HPLC to fractionate the peptides, followed by nano-liquid chromatography-tandem MS (LC-MS/MS) or direct infusion electrospray ionization-(ESI)-MS/MS or MALDI-MS/MS analyses. We detected marked differences in the venom peptidomes and identified peptides ranging from 7 to 39 residues in length by de novo sequencing. Forty-four unique sequences were manually identified, out of which 30 are new peptides, including 17 bradykinin-potentiating peptides, three poly-histidine-poly-glycine peptides and interestingly, 10 L-amino acid oxidase fragments. Some of the new bradykinin-potentiating peptides display significant bradykinin potentiating activity. Automated database search revealed fragments from several toxins in the peptidomes, mainly from l-amino acid oxidase, and allowed the determination of the peptide bond specificity of proteinases and amino acid occurrences for the P4-P4' sites. We also demonstrate that the venom lyophilization/resolubilization process greatly increases the complexity of the peptidome because of the imbalance caused to the venom proteome and the consequent activity of proteinases on venom components. The use of proteinase inhibitors clearly showed different outcomes in the peptidome characterization and suggested that degradomic-peptidomic analysis of snake venoms is highly sensitive to the conditions of sampling procedures.
Figures




References
-
- Warshawsky H., Haddad A., Goncalves R. P., Valeri V., De Lucca F. L. (1973) Fine structure of the venom gland epithelium of the South American rattlesnake and radioautographic studies of protein formation by the secretory cells. Am. J. Anat. 138, 79–119 - PubMed
-
- Junqueira-de-Azevedo I. L., Ho P. L. (2002) A survey of gene expression and diversity in the venom glands of the pitviper snake Bothrops insularis through the generation of expressed sequence tags (ESTs). Gene 299, 279–291 - PubMed
-
- Odell G., Ferry P., Vick L., Fenton A., Decker L., Cowell R., Ownby C., Gutierrez J. (1998) Citrate inhibition of snake venom proteases. Toxicon 36, 1801–1806 - PubMed
-
- Robeva A., Politi V., Shannon J., Bjarnason J., Fox J. (1991) Synthetic and endogenous inhibitors of snake venom metalloproteinases. Biomed. Biochim. Acta 50, 769–773 - PubMed
-
- Serrano S., Shannon J., Wang D., Camargo A., Fox J. (2005) A multifaceted analysis of viperid snake venoms by two-dimensional gel electrophoresis: an approach to understanding venom proteomics. Proteomics 5, 501–510 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

