Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice
- PMID: 22869596
- PMCID: PMC3413350
- DOI: 10.1083/jcb.201204098
Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice
Abstract
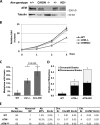
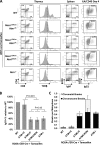
Ataxia telangiectasia (A-T) mutated (ATM) kinase orchestrates deoxyribonucleic acid (DNA) damage responses by phosphorylating numerous substrates implicated in DNA repair and cell cycle checkpoint activation. A-T patients and mouse models that express no ATM protein undergo normal embryonic development but exhibit pleiotropic DNA repair defects. In this paper, we report that mice carrying homozygous kinase-dead mutations in Atm (Atm(KD/KD)) died during early embryonic development. Atm(KD/-) cells exhibited proliferation defects and genomic instability, especially chromatid breaks, at levels higher than Atm(-/-) cells. Despite this increased genomic instability, Atm(KD/-) lymphocytes progressed through variable, diversity, and joining recombination and immunoglobulin class switch recombination, two events requiring nonhomologous end joining, at levels comparable to Atm(-/-) lymphocytes. Together, these results reveal an essential function of ATM during embryogenesis and an important function of catalytically inactive ATM protein in DNA repair.
Figures


Comment in
-
The ATM protein: the importance of being active.J Cell Biol. 2012 Aug 6;198(3):273-5. doi: 10.1083/jcb.201207063. J Cell Biol. 2012. PMID: 22869592 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

