Interpreting the widespread nonlinear force spectra of intermolecular bonds
- PMID: 22869712
- PMCID: PMC3427124
- DOI: 10.1073/pnas.1202946109
Interpreting the widespread nonlinear force spectra of intermolecular bonds
Erratum in
- Proc Natl Acad Sci U S A. 2013 Mar 12;110(11):4429
Abstract
Single molecule force spectroscopy probes the strength, lifetime, and energetic details of intermolecular interactions in a simple experiment. A growing number of these studies have reported distinctly nonlinear trends in rupture force with loading rate that are typically explained in conventional models by invoking complex escape pathways. Recent analyses suggested that these trends should be expected even for simple barriers based on the basic assumptions of bond rupture dynamics and thus may represent the norm rather than the exception. Here we explore how these nonlinear trends reflect the two fundamental regimes of bond rupture: (i) a near-equilibrium regime, produced either by bond reforming in the case of a single bond or by asynchronized rupture of multiple individual bonds, and (ii) a kinetic regime produced by fast, non-equilibrium bond rupture. We analyze both single- and multi-bonded cases, describe the full evolution of the system as it transitions between near- and far-from-equilibrium loading regimes, and show that both interpretations produce essentially identical force spectra. Data from 10 different molecular systems show that this model provides a comprehensive description of force spectra for a diverse suite of bonds over experimentally relevant loading rates, removes the inconsistencies of previous interpretations of transition state distances, and gives ready access to both kinetic and thermodynamic information about the interaction. These results imply that single-molecule binding free energies for a vast number of bonds have already been measured.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
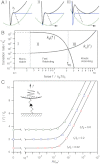
 , xt = 1 Å, ΔGbu = 10 kBT, and kc = 100 pN/nm. Rates are in units of
, xt = 1 Å, ΔGbu = 10 kBT, and kc = 100 pN/nm. Rates are in units of  and force is in units of kBT/xt. The regimes corresponding to stages in the upper panels are partitioned for comparison. (C) The differential equation defining the two-state master equation [Eq. 1] is solved by numerical integration (symbols) with the Bulirsch-Stoer method using Richardson extrapolation (Igor Pro, Wavemetrics). Results are shown for ΔGbu = 10 kBT, and three different confinement force values fk = kcxt, which govern the effects of the probing spring constant on the resulting spectrum. The single-bond model in Eq. 6 is calculated with identical parameters for comparison (solid curves).
and force is in units of kBT/xt. The regimes corresponding to stages in the upper panels are partitioned for comparison. (C) The differential equation defining the two-state master equation [Eq. 1] is solved by numerical integration (symbols) with the Bulirsch-Stoer method using Richardson extrapolation (Igor Pro, Wavemetrics). Results are shown for ΔGbu = 10 kBT, and three different confinement force values fk = kcxt, which govern the effects of the probing spring constant on the resulting spectrum. The single-bond model in Eq. 6 is calculated with identical parameters for comparison (solid curves).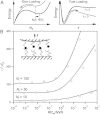
 . Rupture forces for a given trajectory are determined when Nb = 0, and hundreds of trajectories are used to produce the mean rupture force (symbols) plotted here against the dimensionless loading rate. For comparison, the multi-bond model in Eq. 9 is calculated (solid curves) for the same ratio of transition rates, and with
. Rupture forces for a given trajectory are determined when Nb = 0, and hundreds of trajectories are used to produce the mean rupture force (symbols) plotted here against the dimensionless loading rate. For comparison, the multi-bond model in Eq. 9 is calculated (solid curves) for the same ratio of transition rates, and with  .
.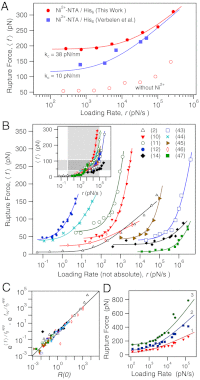
 are shared. Fitted values are xt = 0.78 Å,
are shared. Fitted values are xt = 0.78 Å,  , and feq = 24.6 pN (N = 1), 58.5 pN (N = 2), 142.3 pN (N = 3). Legend in (B) refers to references and corresponding bonds as follows: Biotin/Avidin (2); LFA-1/ICAM-1 [rest 3A9] (10); Aβ-40/Aβ-40 (11); N,C,N-pincer/pyridine (12); Si3N4/Mica in Ethanol (14); peptide/steel (43); Integrin/Fibronectin (; Lysozyme/Anti-Lysozyme (45); Dig/Anti-Dig (46); Actomyosin/ADP (47).
, and feq = 24.6 pN (N = 1), 58.5 pN (N = 2), 142.3 pN (N = 3). Legend in (B) refers to references and corresponding bonds as follows: Biotin/Avidin (2); LFA-1/ICAM-1 [rest 3A9] (10); Aβ-40/Aβ-40 (11); N,C,N-pincer/pyridine (12); Si3N4/Mica in Ethanol (14); peptide/steel (43); Integrin/Fibronectin (; Lysozyme/Anti-Lysozyme (45); Dig/Anti-Dig (46); Actomyosin/ADP (47).References
-
- Evans E. Energy landscapes of biomolecular adhesion and receptor anchoring at interfaces explored with dynamic force spectroscopy. Faraday Discuss. 1998;111:1–16. - PubMed
-
- Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. - PubMed
-
- Eyring H. Viscosity, plasticity, and diffusion as examples of absolute reaction rates. J Chem Phys. 1936;4:283–291.
-
- Tobolsky A, Eyring H. Mechanical properties of polymeric materials. J Chem Phys. 1943;11:125–134.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

