Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase
- PMID: 22869737
- PMCID: PMC3427106
- DOI: 10.1073/pnas.1206187109
Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase
Abstract
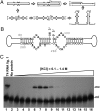
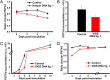
Viroids are a unique class of noncoding RNAs: composed of only a circular, single-stranded molecule of 246-401 nt, they manage to replicate, move, circumvent host defenses, and frequently induce disease in higher plants. Viroids replicate through an RNA-to-RNA rolling-circle mechanism consisting of transcription of oligomeric viroid RNA intermediates, cleavage to unit-length strands, and circularization. Though the host RNA polymerase II (redirected to accept RNA templates) mediates RNA synthesis and a type-III RNase presumably cleavage of Potato spindle tuber viroid (PSTVd) and closely related members of the family Pospiviroidae, the host enzyme catalyzing the final circularization step, has remained elusive. In this study we propose that PSTVd subverts host DNA ligase 1, converting it to an RNA ligase, for the final step. To support this hypothesis, we show that the tomato (Solanum lycopersicum L.) DNA ligase 1 specifically and efficiently catalyzes circularization of the genuine PSTVd monomeric linear replication intermediate opened at position G95-G96 and containing 5'-phosphomonoester and 3'-hydroxyl terminal groups. Moreover, we also show a decreased PSTVd accumulation and a reduced ratio of monomeric circular to total monomeric PSTVd forms in Nicotiana benthamiana Domin plants in which the endogenous DNA ligase 1 was silenced. Thus, in a remarkable example of parasitic strategy, viroids reprogram for their replication the template and substrate specificity of a DNA-dependent RNA polymerase and a DNA ligase to act as RNA-dependent RNA polymerase and RNA ligase, respectively.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Diener TO, Raymer WB. Potato spindle tuber virus: A plant virus with properties of a free nucleic acid. Science. 1967;158:378–381. - PubMed
-
- Gross HJ, et al. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978;273:203–208. - PubMed
-
- Semancik JS, Weathers LG. Exocortis virus of citrus: Association of infectivity with nucleic acid preparations. Virology. 1968;36:326–328. - PubMed
-
- Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. - PubMed
-
- Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol. 2009;47:105–131. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

