Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing
- PMID: 22870198
- PMCID: PMC3411578
- DOI: 10.1371/journal.pone.0040409
Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing
Abstract
Campylobacter jejuni is the most common cause of human bacterial gastroenteritis and is associated with several post-infectious manifestations, including onset of the autoimmune neuropathy Guillain-Barré syndrome, causing significant morbidity and mortality. Poorly-cooked chicken meat is the most frequent source of infection as C. jejuni colonizes the avian intestine in a commensal relationship. However, not all chickens are equally colonized and resistance seems to be genetically determined. We hypothesize that differences in immune response may contribute to variation in colonization levels between susceptible and resistant birds. Using high-throughput sequencing in an avian infection model, we investigate gene expression associated with resistance or susceptibility to colonization of the gastrointestinal tract with C. jejuni and find that gut related immune mechanisms are critical for regulating colonization. Amongst a single population of 300 4-week old chickens, there was clear segregation in levels of C. jejuni colonization 48 hours post-exposure. RNAseq analysis of caecal tissue from 14 C. jejuni-susceptible and 14 C. jejuni-resistant birds generated over 363 million short mRNA sequences which were investigated to identify 219 differentially expressed genes. Significantly higher expression of genes involved in the innate immune response, cytokine signaling, B cell and T cell activation and immunoglobulin production, as well as the renin-angiotensin system was observed in resistant birds, suggesting an early active immune response to C. jejuni. Lower expression of these genes in colonized birds suggests suppression or inhibition of a clearing immune response thus facilitating commensal colonization and generating vectors for zoonotic transmission. This study describes biological processes regulating C. jejuni colonization of the avian intestine and gives insight into the differential immune mechanisms incited in response to commensal bacteria in general within vertebrate populations. The results reported here illustrate how an exaggerated immune response may be elicited in a subset of the population, which alters host-microbe interactions and inhibits the commensal state, therefore having wider relevance with regard to inflammatory and autoimmune disease.
Conflict of interest statement
Figures
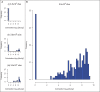
 ), medium (ii, 3.5×10
), medium (ii, 3.5×10 ) and high (iii, 3.5×10
) and high (iii, 3.5×10 ) doses of C. jejuni, and the colonization status of their caeca estimated after 48 hours. No colonization was detected in any bird after infection with 3.5×10
) doses of C. jejuni, and the colonization status of their caeca estimated after 48 hours. No colonization was detected in any bird after infection with 3.5×10 CFU. All birds were colonized after high dosage. Maximum differentiation is achieved after inoculation with the medium dose of 3.5×10
CFU. All birds were colonized after high dosage. Maximum differentiation is achieved after inoculation with the medium dose of 3.5×10 CFU C. jejuni. (B) 255 birds were challenged with 3.5×10
CFU C. jejuni. (B) 255 birds were challenged with 3.5×10 C. jejuni and their caecal colonization status determined after 48 hours. No C. jejuni colonization could be detected in 38 birds. Colonization levels of the remaining birds varied from 2×10
C. jejuni and their caecal colonization status determined after 48 hours. No C. jejuni colonization could be detected in 38 birds. Colonization levels of the remaining birds varied from 2×10 – 4 ×10
– 4 ×10 CFU/g with the majority of this group having very high caecal C. jejuni levels (
CFU/g with the majority of this group having very high caecal C. jejuni levels ( 10
10 CFU/g C. jejuni).
CFU/g C. jejuni).
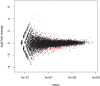
 0.01 are highlighted red.
0.01 are highlighted red.
Similar articles
-
Transplanted human fecal microbiota enhanced Guillain Barré syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice.Microbiome. 2017 Aug 8;5(1):92. doi: 10.1186/s40168-017-0284-4. Microbiome. 2017. PMID: 28789710 Free PMC article.
-
A novel mouse model of Campylobacter jejuni gastroenteritis reveals key pro-inflammatory and tissue protective roles for Toll-like receptor signaling during infection.PLoS Pathog. 2014 Jul 17;10(7):e1004264. doi: 10.1371/journal.ppat.1004264. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25033044 Free PMC article.
-
The immunobiology of Campylobacter jejuni: Innate immunity and autoimmune diseases.Immunobiology. 2016 Apr;221(4):535-43. doi: 10.1016/j.imbio.2015.12.005. Epub 2015 Dec 8. Immunobiology. 2016. PMID: 26709064 Review.
-
Intestinal colonization and acute immune response in commercial turkeys following inoculation with Campylobacter jejuni constructs encoding antibiotic-resistance markers.Vet Immunol Immunopathol. 2019 Apr;210:6-14. doi: 10.1016/j.vetimm.2019.02.003. Epub 2019 Mar 1. Vet Immunol Immunopathol. 2019. PMID: 30947981
-
A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut.Crit Rev Microbiol. 2012 Feb;38(1):17-29. doi: 10.3109/1040841X.2011.615298. Epub 2011 Oct 13. Crit Rev Microbiol. 2012. PMID: 21995731 Review.
Cited by
-
Controlled Intestinal Microbiota Colonisation in Broilers under the Industrial Production System.Animals (Basel). 2022 Nov 25;12(23):3296. doi: 10.3390/ani12233296. Animals (Basel). 2022. PMID: 36496817 Free PMC article.
-
Colonization of a commercial broiler line by Campylobacter is under limited genetic control and does not significantly impair performance or intestinal health.Poult Sci. 2018 Dec 1;97(12):4167-4176. doi: 10.3382/ps/pey295. Poult Sci. 2018. PMID: 29982748 Free PMC article.
-
Highly multiplexed quantitative PCR-based platform for evaluation of chicken immune responses.PLoS One. 2019 Dec 3;14(12):e0225658. doi: 10.1371/journal.pone.0225658. eCollection 2019. PLoS One. 2019. PMID: 31794562 Free PMC article.
-
Quantitative trait loci and transcriptome signatures associated with avian heritable resistance to Campylobacter.Sci Rep. 2021 Jan 12;11(1):1623. doi: 10.1038/s41598-020-79005-7. Sci Rep. 2021. PMID: 33436657 Free PMC article.
-
N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness.Infect Immun. 2013 May;81(5):1674-82. doi: 10.1128/IAI.01370-12. Epub 2013 Mar 4. Infect Immun. 2013. PMID: 23460522 Free PMC article.
References
-
- Hooper LV, Macpherson AJ (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10: 159–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

