Genotype×age interaction in human transcriptional ageing
- PMID: 22871458
- PMCID: PMC3541784
- DOI: 10.1016/j.mad.2012.07.005
Genotype×age interaction in human transcriptional ageing
Abstract
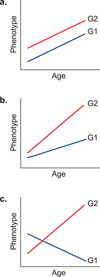
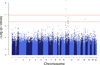
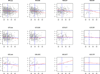
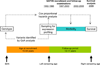
Individual differences in biological ageing (i.e., the rate of physiological response to the passage of time) may be due in part to genotype-specific variation in gene action. However, the sources of heritable variation in human age-related gene expression profiles are largely unknown. We have profiled genome-wide expression in peripheral blood mononuclear cells from 1240 individuals in large families and found 4472 human autosomal transcripts, representing ~4349 genes, significantly correlated with age. We identified 623 transcripts that show genotype by age interaction in addition to a main effect of age, defining a large set of novel candidates for characterization of the mechanisms of differential biological ageing. We applied a novel SNP genotype × age interaction test to one of these candidates, the ubiquilin-like gene UBQLNL, and found evidence of joint cis-association and genotype by age interaction as well as trans-genotype by age interaction for UBQLNL expression. Both UBQLNL expression levels at recruitment and cis genotype are associated with longitudinal cancer risk in our study cohort.
Copyright © 2012 Elsevier Ireland Ltd. All rights reserved.
Figures




References
-
- Almasy L, Towne B, Peterson C, Blangero J. Detecting genotype×age interaction. Genet. Epidemiol. 2001;21(suppl. 1):S819–S824. - PubMed
-
- Arking R. The Biology of Aging: Observations and Principles. 3rd ed. New York: Oxford University Press; 2006.
-
- Bell G. The ecology and genetics of fitness inChlamydomonas I. Genotype-by-environment interaction among pure strains. Proc. R. Soc. Lond. B. 1990;240(1298):295–321.
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B. 1995;57(1):289–300.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

