Anterior-posterior differences in HoxD chromatin topology in limb development
- PMID: 22872084
- PMCID: PMC3413162
- DOI: 10.1242/dev.081174
Anterior-posterior differences in HoxD chromatin topology in limb development
Abstract
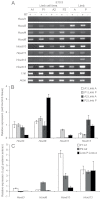
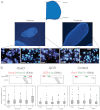
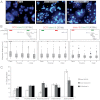
A late phase of HoxD activation is crucial for the patterning and growth of distal structures across the anterior-posterior (A-P) limb axis of mammals. Polycomb complexes and chromatin compaction have been shown to regulate Hox loci along the main body axis in embryonic development, but the extent to which they have a role in limb-specific HoxD expression, an evolutionary adaptation defined by the activity of distal enhancer elements that drive expression of 5' Hoxd genes, has yet to be fully elucidated. We reveal two levels of chromatin topology that differentiate distal limb A-P HoxD activity. Using both immortalised cell lines derived from posterior and anterior regions of distal E10.5 mouse limb buds, and analysis in E10.5 dissected limb buds themselves, we show that there is a loss of polycomb-catalysed H3K27me3 histone modification and a chromatin decompaction over HoxD in the distal posterior limb compared with anterior. Moreover, we show that the global control region (GCR) long-range enhancer spatially colocalises with the 5' HoxD genomic region specifically in the distal posterior limb. This is consistent with the formation of a chromatin loop between 5' HoxD and the GCR regulatory module at the time and place of distal limb bud development when the GCR participates in initiating Hoxd gene quantitative collinearity and Hoxd13 expression. This is the first example of A-P differences in chromatin compaction and chromatin looping in the development of the mammalian secondary body axis (limb).
Figures




References
-
- Amano T., Sagai T., Tanabe H., Mizushina Y., Nakazawa H., Shiroishi T. (2009). Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. B 57, 289–300
-
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nat. Cell Biol. 441, 349–353 - PubMed
-
- Chambeyron S., Da, Silva N. R., Lawson K. A., Bickmore W. A. (2005). Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132, 2215–2223 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

