DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs
- PMID: 22872150
- PMCID: PMC3442272
- DOI: 10.1038/emboj.2012.220
DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs
Abstract
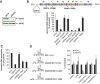
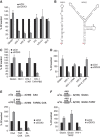
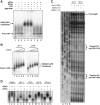
Here, we have characterized a step in translation initiation of viral and cellular mRNAs that contain RNA secondary structures immediately at the vicinity of their m(7)GTP cap. This is mediated by the DEAD-box helicase DDX3 which can directly bind to the 5' of the target mRNA where it clamps the entry of eIF4F through an eIF4G and Poly A-binding protein cytoplasmic 1 (PABP) double interaction. This could induce limited local strand separation of the secondary structure to allow 43S pre-initiation complex attachment to the 5' free extremity of the mRNA. We further demonstrate that the requirement for DDX3 is highly specific to some selected transcripts, cannot be replaced or substituted by eIF4A and is only needed in the very early steps of ribosome binding and prior to 43S ribosomal scanning. Altogether, these data define an unprecedented role for a DEAD-box RNA helicase in translation initiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
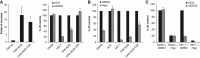
References
-
- Bannwarth S, Gatignol A (2005) HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res 3: 61–71 - PubMed
-
- Banroques J, Doere M, Dreyfus M, Linder P, Tanner NK (2010) Motif III in superfamily 2 ‘helicases’ helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol 396: 949–966 - PubMed
-
- Berkhout B (2000) Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv Pharmacol 48: 29–73 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

