Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation
- PMID: 22872859
- PMCID: PMC3494204
- DOI: 10.1074/mcp.M112.017194
Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation
Abstract
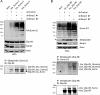
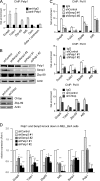
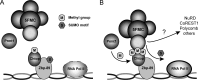
Chromatin target of Prmt1 (Chtop) is a vertebrate-specific chromatin-bound protein that plays an important role in transcriptional regulation. As its mechanism of action remains unclear, we identified Chtop-interacting proteins using a biotinylation-proteomics approach. Here we describe the identification and initial characterization of Five Friends of Methylated Chtop (5FMC). 5FMC is a nuclear complex that can only be recruited by Chtop when the latter is arginine-methylated by Prmt1. It consists of the co-activator Pelp1, the Sumo-specific protease Senp3, Wdr18, Tex10, and Las1L. Pelp1 functions as the core of 5FMC, as the other components become unstable in the absence of Pelp1. We show that recruitment of 5FMC to Zbp-89, a zinc-finger transcription factor, affects its sumoylation status and transactivation potential. Collectively, our data provide a mechanistic link between arginine methylation and (de)sumoylation in the control of transcriptional activity.
Figures




References
-
- O'Brien K. B., Alberich-Jordà M., Yadav N., Kocher O., Diruscio A., Ebralidze A., Levantini E., Sng N. J., Bhasin M., Caron T., Kim D., Steidl U., Huang G., Halmos B., Rodig S. J., Bedford M. T., Tenen D. G., Kobayashi S. (2010) CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 137, 2147–2156 - PMC - PubMed
-
- Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., Allis C. D., Tempst P., Nimer S. D. (2008) Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22, 640–653 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

