Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis
- PMID: 22873208
- PMCID: PMC3425129
- DOI: 10.1186/1741-7007-10-71
Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis
Abstract
Background: Membrane-bound organelles are a defining feature of eukaryotic cells, and play a central role in most of their fundamental processes. The Rab G proteins are the single largest family of proteins that participate in the traffic between organelles, with 66 Rabs encoded in the human genome. Rabs direct the organelle-specific recruitment of vesicle tethering factors, motor proteins, and regulators of membrane traffic. Each organelle or vesicle class is typically associated with one or more Rab, with the Rabs present in a particular cell reflecting that cell's complement of organelles and trafficking routes.
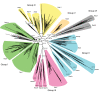
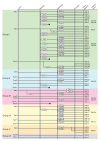
Results: Through iterative use of hidden Markov models and tree building, we classified Rabs across the eukaryotic kingdom to provide the most comprehensive view of Rab evolution obtained to date. A strikingly large repertoire of at least 20 Rabs appears to have been present in the last eukaryotic common ancestor (LECA), consistent with the 'complexity early' view of eukaryotic evolution. We were able to place these Rabs into six supergroups, giving a deep view into eukaryotic prehistory.
Conclusions: Tracing the fate of the LECA Rabs revealed extensive losses with many extant eukaryotes having fewer Rabs, and none having the full complement. We found that other Rabs have expanded and diversified, including a large expansion at the dawn of metazoans, which could be followed to provide an account of the evolutionary history of all human Rabs. Some Rab changes could be correlated with differences in cellular organization, and the relative lack of variation in other families of membrane-traffic proteins suggests that it is the changes in Rabs that primarily underlies the variation in organelles between species and cell types.
Figures



Comment in
-
The Rabs: a family at the root of metazoan evolution.BMC Biol. 2012 Aug 8;10:68. doi: 10.1186/1741-7007-10-68. BMC Biol. 2012. PMID: 22873178 Free PMC article.
References
-
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. - PubMed
-
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. - PubMed
-
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

