A Sox9/Fgf feed-forward loop maintains pancreatic organ identity
- PMID: 22874919
- PMCID: PMC3424044
- DOI: 10.1242/dev.078733
A Sox9/Fgf feed-forward loop maintains pancreatic organ identity
Abstract
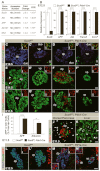
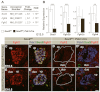
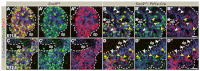
All mature pancreatic cell types arise from organ-specific multipotent progenitor cells. Although previous studies have identified cell-intrinsic and -extrinsic cues for progenitor cell expansion, it is unclear how these cues are integrated within the niche of the developing organ. Here, we present genetic evidence in mice that the transcription factor Sox9 forms the centerpiece of a gene regulatory network that is crucial for proper organ growth and maintenance of organ identity. We show that pancreatic progenitor-specific ablation of Sox9 during early pancreas development causes pancreas-to-liver cell fate conversion. Sox9 deficiency results in cell-autonomous loss of the fibroblast growth factor receptor (Fgfr) 2b, which is required for transducing mesenchymal Fgf10 signals. Likewise, Fgf10 is required to maintain expression of Sox9 and Fgfr2 in epithelial progenitors, showing that Sox9, Fgfr2 and Fgf10 form a feed-forward expression loop in the early pancreatic organ niche. Mirroring Sox9 deficiency, perturbation of Fgfr signaling in pancreatic explants or genetic inactivation of Fgf10 also result in hepatic cell fate conversion. Combined with previous findings that Fgfr2b or Fgf10 are necessary for pancreatic progenitor cell proliferation, our results demonstrate that organ fate commitment and progenitor cell expansion are coordinately controlled by the activity of a Sox9/Fgf10/Fgfr2b feed-forward loop in the pancreatic niche. This self-promoting Sox9/Fgf10/Fgfr2b loop may regulate cell identity and organ size in a broad spectrum of developmental and regenerative contexts.
Figures


Similar articles
-
A branching morphogenesis program governs embryonic growth of the thyroid gland.Development. 2018 Jan 25;145(2):dev146829. doi: 10.1242/dev.146829. Development. 2018. PMID: 29361553 Free PMC article.
-
Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via beta-catenin activation.Hepatology. 2007 Oct;46(4):1187-97. doi: 10.1002/hep.21814. Hepatology. 2007. PMID: 17668871 Free PMC article.
-
Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells.Dev Dyn. 2003 Oct;228(2):185-93. doi: 10.1002/dvdy.10368. Dev Dyn. 2003. PMID: 14517990
-
The role of SOX9 transcription factor in pancreatic and duodenal development.Stem Cells Dev. 2013 Nov 15;22(22):2935-43. doi: 10.1089/scd.2013.0106. Epub 2013 Aug 2. Stem Cells Dev. 2013. PMID: 23806070 Review.
-
FGF10: A multifunctional mesenchymal-epithelial signaling growth factor in development, health, and disease.Cytokine Growth Factor Rev. 2016 Apr;28:63-9. doi: 10.1016/j.cytogfr.2015.10.001. Epub 2015 Oct 31. Cytokine Growth Factor Rev. 2016. PMID: 26559461 Review.
Cited by
-
Dissecting FGF Signalling to Target Cellular Crosstalk in Pancreatic Cancer.Cells. 2021 Apr 8;10(4):847. doi: 10.3390/cells10040847. Cells. 2021. PMID: 33918004 Free PMC article. Review.
-
Role of FGF10/FGFR2b Signaling in Mouse Digestive Tract Development, Repair and Regeneration Following Injury.Front Cell Dev Biol. 2019 Dec 10;7:326. doi: 10.3389/fcell.2019.00326. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31921841 Free PMC article. Review.
-
Lung epithelial branching program antagonizes alveolar differentiation.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):18042-51. doi: 10.1073/pnas.1311760110. Epub 2013 Sep 20. Proc Natl Acad Sci U S A. 2013. PMID: 24058167 Free PMC article.
-
Pancreas morphogenesis and homeostasis depends on tightly regulated Zeb1 levels in epithelial cells.Cell Death Discov. 2021 Jun 11;7(1):138. doi: 10.1038/s41420-021-00522-z. Cell Death Discov. 2021. PMID: 34112759 Free PMC article.
-
Human stem cell models: lessons for pancreatic development and disease.Genes Dev. 2019 Nov 1;33(21-22):1475-1490. doi: 10.1101/gad.331397.119. Genes Dev. 2019. PMID: 31676735 Free PMC article. Review.
References
-
- Ahlgren U., Jonsson J., Edlund H. (1996). The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development 122, 1409–1416 - PubMed
-
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D. J., Honjo T., Hrabe de Angelis M., Lendahl U., Edlund H. (1999). Notch signalling controls pancreatic cell differentiation. Nature 400, 877–881 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

