Identification and complete genome characterization of a novel picornavirus in turkey (Meleagris gallopavo)
- PMID: 22875254
- PMCID: PMC3541788
- DOI: 10.1099/vir.0.043224-0
Identification and complete genome characterization of a novel picornavirus in turkey (Meleagris gallopavo)
Abstract
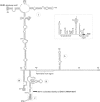
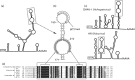
Members of the family Picornaviridae are important pathogens of humans and animals, although compared with the thousands of known bird species (>10 000), only a few (n = 11) picornaviruses have been identified from avian sources. This study reports the metagenomic detection and complete genome characterization of a novel turkey picornavirus from faecal samples collected from eight turkey farms in Hungary. Using RT-PCR, both healthy (two of three) and affected (seven of eight) commercial turkeys with enteric and/or stunting syndrome were shown to be shedding viruses in seven (88 %) of the eight farms. The viral genome sequence (turkey/M176/2011/HUN; GenBank accession no. JQ691613) shows a high degree of amino acid sequence identity (96 %) to the partial P3 genome region of a picornavirus reported recently in turkey and chickens from the USA and probably belongs to the same species. In the P1 and P2 regions, turkey/M176/2011/HUN is related most closely to, but distinct from, the kobuviruses and turdivirus 1. Complete genome analysis revealed the presence of characteristic picornaviral amino acid motifs, a potential type II-like 5' UTR internal ribosome entry site (first identified among avian-origin picornaviruses) and a conserved, 48 nt long 'barbell-like' structure found at the 3' UTR of turkey/M176/2011/HUN and members of the picornavirus genera Avihepatovirus and Kobuvirus. The general presence of turkey picornavirus - a novel picornavirus species - in faecal samples from healthy and affected turkeys in Hungary and in the USA suggests the worldwide occurrence and endemic circulation of this virus in turkey farms. Further studies are needed to investigate the aetiological role and pathogenic potential of this picornavirus in food animals.
Figures




References
-
- Alexandersen S., Knowles N. J., Dekker A., Belsham G. J., Zhang Z., Koenen F. (2012). Picornaviruses. In Diseases of Swine, 10th edn, pp. 587–620 Edited by Zimmerman J. J., Karriker L. A., Ramirez A., Schwartz K. J., Stevenson G. W. Chichester, UK: Wiley
-
- Bailey D., Karakasiliotis I., Vashist S., Chung L. M., Rees J., McFadden N., Benson A., Yarovinsky F., Simmonds P., Goodfellow I. (2010). Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J Virol 84, 2859–2870 10.1128/JVI.02053-09 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

