Cross-platform pathway-based analysis identifies markers of response to the PARP inhibitor olaparib
- PMID: 22875744
- PMCID: PMC3429780
- DOI: 10.1007/s10549-012-2188-0
Cross-platform pathway-based analysis identifies markers of response to the PARP inhibitor olaparib
Abstract
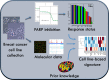
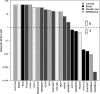
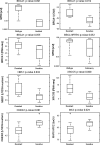
Poly(ADP-ribose) polymerase (PARP) is an enzyme involved in DNA repair. PARP inhibitors can act as chemosensitizers, or operate on the principle of synthetic lethality when used as single agent. Clinical trials have shown drugs in this class to be promising for BRCA mutation carriers. We postulated that inability to demonstrate response in non-BRCA carriers in which BRCA is inactivated by other mechanisms or with deficiency in homologous recombination for DNA repair is due to lack of molecular markers that define a responding subpopulation. We identified candidate markers for this purpose for olaparib (AstraZeneca) by measuring inhibitory effects of nine concentrations of olaparib in 22 breast cancer cell lines and identifying features in transcriptional and genome copy number profiles that were significantly correlated with response. We emphasized in this discovery process genes involved in DNA repair. We found that the cell lines that were sensitive to olaparib had a significant lower copy number of BRCA1 compared to the resistant cell lines (p value 0.012). In addition, we discovered seven genes from DNA repair pathways whose transcriptional levels were associated with response. These included five genes (BRCA1, MRE11A, NBS1, TDG, and XPA) whose transcript levels were associated with resistance and two genes (CHEK2 and MK2) whose transcript levels were associated with sensitivity. We developed an algorithm to predict response using the seven-gene transcription levels and applied it to 1,846 invasive breast cancer samples from 8 U133A/plus 2 (Affymetrix) data sets and found that 8-21 % of patients would be predicted to be responsive to olaparib. A similar response frequency was predicted in 536 samples analyzed on an Agilent platform. Importantly, tumors predicted to respond were enriched in basal subtype tumors. Our studies support clinical evaluation of the utility of our seven-gene signature as a predictor of response to olaparib.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

