Quiescent fibroblasts are protected from proteasome inhibition-mediated toxicity
- PMID: 22875985
- PMCID: PMC3442405
- DOI: 10.1091/mbc.E12-03-0192
Quiescent fibroblasts are protected from proteasome inhibition-mediated toxicity
Abstract
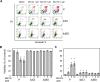
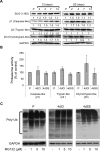
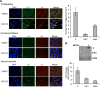
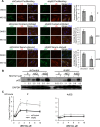
Proteasome inhibition is used as a treatment strategy for multiple types of cancers. Although proteasome inhibition can induce apoptotic cell death in actively proliferating cells, it is less effective in quiescent cells. In this study, we used primary human fibroblasts as a model system to explore the link between the proliferative state of a cell and proteasome inhibition-mediated cell death. We found that proliferating and quiescent fibroblasts have strikingly different responses to MG132, a proteasome inhibitor; proliferating cells rapidly apoptosed, whereas quiescent cells maintained viability. Moreover, MG132 treatment of proliferating fibroblasts led to increased superoxide anion levels, juxtanuclear accumulation of ubiquitin- and p62/SQSTM1-positive protein aggregates, and apoptotic cell death, whereas MG132-treated quiescent cells displayed fewer juxtanuclear protein aggregates, less apoptosis, and higher levels of mitochondrial superoxide dismutase. In both cell states, reducing reactive oxygen species with N-acetylcysteine lessened protein aggregation and decreased apoptosis, suggesting that protein aggregation promotes apoptosis. In contrast, increasing cellular superoxide levels with 2-methoxyestradiol treatment or inhibition of autophagy/lysosomal pathways with bafilomycin A1 sensitized serum-starved quiescent cells to MG132-induced apoptosis. Thus, antioxidant defenses and the autophagy/lysosomal pathway protect serum-starved quiescent fibroblasts from proteasome inhibition-induced cytotoxicity.
Figures



References
-
- Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. - PubMed
-
- Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. - PubMed
-
- Cao Y, Klionsky DJ. Physiological functions of Atg6/beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

