Hypothesis-based analysis of gene-gene interactions and risk of myocardial infarction
- PMID: 22876292
- PMCID: PMC3410908
- DOI: 10.1371/journal.pone.0041730
Hypothesis-based analysis of gene-gene interactions and risk of myocardial infarction
Abstract
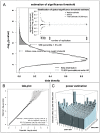
The genetic loci that have been found by genome-wide association studies to modulate risk of coronary heart disease explain only a fraction of its total variance, and gene-gene interactions have been proposed as a potential source of the remaining heritability. Given the potentially large testing burden, we sought to enrich our search space with real interactions by analyzing variants that may be more likely to interact on the basis of two distinct hypotheses: a biological hypothesis, under which MI risk is modulated by interactions between variants that are known to be relevant for its risk factors; and a statistical hypothesis, under which interacting variants individually show weak marginal association with MI. In a discovery sample of 2,967 cases of early-onset myocardial infarction (MI) and 3,075 controls from the MIGen study, we performed pair-wise SNP interaction testing using a logistic regression framework. Despite having reasonable power to detect interaction effects of plausible magnitudes, we observed no statistically significant evidence of interaction under these hypotheses, and no clear consistency between the top results in our discovery sample and those in a large validation sample of 1,766 cases of coronary heart disease and 2,938 controls from the Wellcome Trust Case-Control Consortium. Our results do not support the existence of strong interaction effects as a common risk factor for MI. Within the scope of the hypotheses we have explored, this study places a modest upper limit on the magnitude that epistatic risk effects are likely to have at the population level (odds ratio for MI risk 1.3-2.0, depending on allele frequency and interaction model).
Conflict of interest statement
Figures

References
-
- World Health Organization (2009) GLOBAL HEALTH RISKS. Mortality and burden of disease attributable to selected major risks. 28–29.
-
- Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, et al. (2002) Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med 252: 247–254. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

