13C NMR reveals no evidence of n-π* interactions in proteins
- PMID: 22876300
- PMCID: PMC3410932
- DOI: 10.1371/journal.pone.0042075
13C NMR reveals no evidence of n-π* interactions in proteins
Abstract
An n = π* interaction between neighboring carbonyl groups has been postulated to stabilize protein structures. Such an interaction would affect the (13)C chemical shielding of the carbonyl groups, whose paramagnetic component is dominated by n = π* and π = π* excitations. Model compound calculations indicate that both the interaction energetics and the chemical shielding of the carbonyl group are instead dominated by a classical dipole-dipole interaction. A set of high-resolution protein structures with associated carbonyl (13)C chemical shift assignments verifies this correlation and provides no evidence for an inter-carbonyl n = π* interaction.
Conflict of interest statement
Figures

 interaction between carbonyl groups. (B) 2D contour plot of carbonyl
interaction between carbonyl groups. (B) 2D contour plot of carbonyl  C chemical shift differences relative to random coil values as a function of the distance (
C chemical shift differences relative to random coil values as a function of the distance ( ) and angle (
) and angle ( ) between carbonyls. A Gaussian smoothing function was applied to the data with
) between carbonyls. A Gaussian smoothing function was applied to the data with  and
and  of 0.3 Å and 1.5°, respectively. A transparency mask based on the density of experimental data (see also Figure S1) is overlaid on the contour plot. Regions lacking experimental data are white. Positive values indicate downfield shifts.
of 0.3 Å and 1.5°, respectively. A transparency mask based on the density of experimental data (see also Figure S1) is overlaid on the contour plot. Regions lacking experimental data are white. Positive values indicate downfield shifts.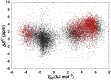
 C chemical shift differences relative to random coil are plotted against calculated dipole-dipole potential (
C chemical shift differences relative to random coil are plotted against calculated dipole-dipole potential ( ). The dipole-dipole potential is calculated from the high-resolution x-ray structure using Equation 1. Pairs of carbonyls with
). The dipole-dipole potential is calculated from the high-resolution x-ray structure using Equation 1. Pairs of carbonyls with  and
and  values within the optimal limits for an
values within the optimal limits for an  interaction are colored red.
interaction are colored red.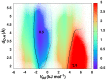
 C carbonyl chemical shift differences as a function of calculated dipole-dipole potential (
C carbonyl chemical shift differences as a function of calculated dipole-dipole potential ( ) and calculated hydrogen bond length (
) and calculated hydrogen bond length ( ). See also Figure S2.
). See also Figure S2.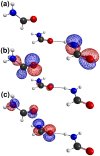
 donor and (C) the putative
donor and (C) the putative  acceptor, in the trimeric complex used in quantum chemical calculations.
acceptor, in the trimeric complex used in quantum chemical calculations.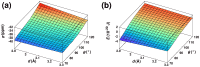
 C chemical shielding (
C chemical shielding ( ) and (B) dipole-dipole interaction energy (
) and (B) dipole-dipole interaction energy ( ) as a function of the distance between donor oxygen and acceptor carbon (
) as a function of the distance between donor oxygen and acceptor carbon ( ) and the angle between carbonyl groups (
) and the angle between carbonyl groups ( ). See also Figure S3.
). See also Figure S3.
 C chemical shielding (
C chemical shielding ( ) as a function of the angle between the carbonyls (
) as a function of the angle between the carbonyls ( ) for three different distances (
) for three different distances ( ) between the donor oxygen and acceptor carbon.
) between the donor oxygen and acceptor carbon.Similar articles
-
13C and 15N NMR studies of iron-bound cyanides of heme proteins and related model complexes: sensitive probe for detecting hydrogen-bonding interactions at the proximal and distal sides.Inorg Chem. 2006 Aug 21;45(17):6816-27. doi: 10.1021/ic0607383. Inorg Chem. 2006. PMID: 16903738
-
The n→π* Interaction.Acc Chem Res. 2017 Aug 15;50(8):1838-1846. doi: 10.1021/acs.accounts.7b00121. Epub 2017 Jul 23. Acc Chem Res. 2017. PMID: 28735540 Free PMC article.
-
Flavin-protein interactions in flavocytochrome b2 as studied by NMR after reconstitution of the enzyme with 13C- and 15N-labelled flavin.Eur J Biochem. 2000 Aug;267(16):5156-67. doi: 10.1046/j.1432-1327.2000.01584.x. Eur J Biochem. 2000. PMID: 10931200
-
Multidimensional solid state NMR of anisotropic interactions in peptides and proteins.J Chem Phys. 2008 Feb 7;128(5):052207. doi: 10.1063/1.2834735. J Chem Phys. 2008. PMID: 18266412 Review.
-
NMR chemical shift data and ab initio shielding calculations: emerging tools for protein structure determination.Chem Soc Rev. 2010 Feb;39(2):578-90. doi: 10.1039/b811366c. Epub 2009 Nov 4. Chem Soc Rev. 2010. PMID: 20111782 Review.
Cited by
-
The Realm of Unconventional Noncovalent Interactions in Proteins: Their Significance in Structure and Function.ACS Omega. 2023 Jun 13;8(25):22268-22284. doi: 10.1021/acsomega.3c00205. eCollection 2023 Jun 27. ACS Omega. 2023. PMID: 37396257 Free PMC article. Review.
-
Secondary Forces in Protein Folding.ACS Chem Biol. 2019 Aug 16;14(8):1677-1686. doi: 10.1021/acschembio.9b00339. Epub 2019 Jun 19. ACS Chem Biol. 2019. PMID: 31243961 Free PMC article. Review.
-
Protein folding, misfolding and aggregation: The importance of two-electron stabilizing interactions.PLoS One. 2017 Sep 18;12(9):e0180905. doi: 10.1371/journal.pone.0180905. eCollection 2017. PLoS One. 2017. PMID: 28922400 Free PMC article.
-
n→π* interactions of amides and thioamides: implications for protein stability.J Am Chem Soc. 2013 May 29;135(21):7843-6. doi: 10.1021/ja4033583. Epub 2013 May 20. J Am Chem Soc. 2013. PMID: 23663100 Free PMC article.
-
Intimate interactions with carbonyl groups: dipole-dipole or n→π*?J Org Chem. 2013 Mar 1;78(5):2099-103. doi: 10.1021/jo302265k. Epub 2012 Dec 10. J Org Chem. 2013. PMID: 23163960 Free PMC article.
References
-
- Rost B (1999) Twilight zone of protein sequence alignments. Protein Engineering 12: 85–94. - PubMed
-
- Robertson AD, Murphy KP (1997) Protein structure and the energetics of protein stability. Chem Rev 97: 1251–1267. - PubMed
-
- Dill KA (1990) Dominant forces in protein folding. Biochemistry 29: 7133–7155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

