High prevalence of Arginine to Glutamine substitution at 98, 141 and 162 positions in Troponin I (TNNI3) associated with hypertrophic cardiomyopathy among Indians
- PMID: 22876777
- PMCID: PMC3495047
- DOI: 10.1186/1471-2350-13-69
High prevalence of Arginine to Glutamine substitution at 98, 141 and 162 positions in Troponin I (TNNI3) associated with hypertrophic cardiomyopathy among Indians
Abstract
Background: Troponin I (TNNI3) is the inhibitory subunit of the thin filament regulatory complex Troponin, which confers calcium-sensitivity to striated muscle actomyosin ATPase activity. Mutations (2-7%) in this gene had been reported in hypertrophic cardiomyopathy patients (HCM). However, the frequencies of mutations and associated clinical presentation have not been established in cardiomyopathy patients of Indian origin, hence we have undertaken this study.
Methods: We have sequenced all the exons, including the exon-intron boundaries of TNNI3 gene in 101 hypertrophic cardiomyopathy patients (HCM), along with 160 healthy controls, inhabited in the same geographical region of southern India.
Results: Our study revealed a total of 16 mutations. Interestingly, we have observed Arginine to Glutamine (R to Q) mutation at 3 positions 98, 141 and 162, exclusively in HCM patients with family history of sudden cardiac death. The novel R98Q was observed in a severe hypertrophic obstructive cardiomyopathy patient (HOCM). The R141Q mutation was observed in two familial cases of severe asymmetric septal hypertrophy (ASH++). The R162Q mutation was observed in a ASH++ patient with mean septal thickness of 29 mm, and have also consists of allelic heterogeneity by means of having one more synonymous (E179E) mutation at g.4797: G → A: in the same exon 7, which replaces a very frequent codon (GAG: 85%) with a rare codon (GAA: 14%). Screening for R162Q mutation in all the available family members revealed its presence in 9 individuals, including 7 with allelic heterogeneity (R162Q and E179E) of which 4 were severely affected. We also found 2 novel SNPs, (g.2653; G → A and g.4003 C → T) exclusively in HCM, and in silico analysis of these SNPs have predicted to cause defect in recognition/binding sites for proteins responsible for proper splicing.
Conclusion: Our study has provided valuable information regarding the prevalence of TNNI3 mutations in Indian HCM patients and its risk assessment, these will help in genetic counseling and to adopt appropriate treatment strategies.
Figures

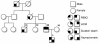
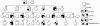
References
-
- Maron BJ. Hypertrophic cardiomyopathy. A systematic review. JAMA. 2002;287:1308–1320. - PubMed
-
- Bhavsar PK, Brand NJ, Yacoub MH, Barton PJR. Isolation and characterization of the human cardiac troponin I gene (TNNI3) Genomics. 1996;35:11–23. - PubMed
-
- Mogensen J, Murphy RT, Kubo T, Bahl A, Moon JC, Klausen IC, Elliott PM, McKenna WJ. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:2315–2325. - PubMed
-
- Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang TH, Choo JA, Chung KS, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–382. - PubMed
-
- Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circul. 2002;105:446–451. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

