AtlasT4SS: a curated database for type IV secretion systems
- PMID: 22876890
- PMCID: PMC3489848
- DOI: 10.1186/1471-2180-12-172
AtlasT4SS: a curated database for type IV secretion systems
Abstract
Background: The type IV secretion system (T4SS) can be classified as a large family of macromolecule transporter systems, divided into three recognized sub-families, according to the well-known functions. The major sub-family is the conjugation system, which allows transfer of genetic material, such as a nucleoprotein, via cell contact among bacteria. Also, the conjugation system can transfer genetic material from bacteria to eukaryotic cells; such is the case with the T-DNA transfer of Agrobacterium tumefaciens to host plant cells. The system of effector protein transport constitutes the second sub-family, and the third one corresponds to the DNA uptake/release system. Genome analyses have revealed numerous T4SS in Bacteria and Archaea. The purpose of this work was to organize, classify, and integrate the T4SS data into a single database, called AtlasT4SS - the first public database devoted exclusively to this prokaryotic secretion system.
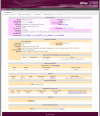
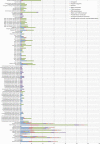
Description: The AtlasT4SS is a manual curated database that describes a large number of proteins related to the type IV secretion system reported so far in Gram-negative and Gram-positive bacteria, as well as in Archaea. The database was created using the RDBMS MySQL and the Catalyst Framework based in the Perl programming language and using the Model-View-Controller (MVC) design pattern for Web. The current version holds a comprehensive collection of 1,617 T4SS proteins from 58 Bacteria (49 Gram-negative and 9 Gram-Positive), one Archaea and 11 plasmids. By applying the bi-directional best hit (BBH) relationship in pairwise genome comparison, it was possible to obtain a core set of 134 clusters of orthologous genes encoding T4SS proteins.
Conclusions: In our database we present one way of classifying orthologous groups of T4SSs in a hierarchical classification scheme with three levels. The first level comprises four classes that are based on the organization of genetic determinants, shared homologies, and evolutionary relationships: (i) F-T4SS, (ii) P-T4SS, (iii) I-T4SS, and (iv) GI-T4SS. The second level designates a specific well-known protein families otherwise an uncharacterized protein family. Finally, in the third level, each protein of an ortholog cluster is classified according to its involvement in a specific cellular process. AtlasT4SS database is open access and is available at http://www.t4ss.lncc.br.
Figures



Similar articles
-
T4SP Database 2.0: An Improved Database for Type IV Secretion Systems in Bacterial Genomes with New Online Analysis Tools.Comput Math Methods Med. 2016;2016:9415459. doi: 10.1155/2016/9415459. Epub 2016 Sep 21. Comput Math Methods Med. 2016. PMID: 27738451 Free PMC article.
-
Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion.Nucleic Acids Res. 2014 May;42(9):5715-27. doi: 10.1093/nar/gku194. Epub 2014 Mar 12. Nucleic Acids Res. 2014. PMID: 24623814 Free PMC article.
-
Membrane and core periplasmic Agrobacterium tumefaciens virulence Type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells.mBio. 2011 Oct 25;2(6):e00218-11. doi: 10.1128/mBio.00218-11. Print 2011. mBio. 2011. PMID: 22027007 Free PMC article.
-
Biological diversity of prokaryotic type IV secretion systems.Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review.
-
The Agrobacterium VirB/VirD4 T4SS: Mechanism and Architecture Defined Through In Vivo Mutagenesis and Chimeric Systems.Curr Top Microbiol Immunol. 2018;418:233-260. doi: 10.1007/82_2018_94. Curr Top Microbiol Immunol. 2018. PMID: 29808338 Free PMC article. Review.
Cited by
-
The expanding bacterial type IV secretion lexicon.Res Microbiol. 2013 Jul-Aug;164(6):620-39. doi: 10.1016/j.resmic.2013.03.012. Epub 2013 Mar 28. Res Microbiol. 2013. PMID: 23542405 Free PMC article. Review.
-
Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms.BMC Genomics. 2014 Jan 22;15:54. doi: 10.1186/1471-2164-15-54. BMC Genomics. 2014. PMID: 24450656 Free PMC article.
-
SecretEPDB: a comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems.Sci Rep. 2017 Jan 23;7:41031. doi: 10.1038/srep41031. Sci Rep. 2017. PMID: 28112271 Free PMC article.
-
traG Gene Is Conserved across Mesorhizobium spp. Able to Nodulate the Same Host Plant and Expressed in Response to Root Exudates.Biomed Res Int. 2019 Jan 30;2019:3715271. doi: 10.1155/2019/3715271. eCollection 2019. Biomed Res Int. 2019. PMID: 30834262 Free PMC article.
-
Comparative genomics of multiple strains of Pseudomonas cannabina pv. alisalensis, a potential model pathogen of both monocots and dicots.PLoS One. 2013;8(3):e59366. doi: 10.1371/journal.pone.0059366. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555661 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

