Loss of the tumor suppressor BAP1 causes myeloid transformation
- PMID: 22878500
- PMCID: PMC5201002
- DOI: 10.1126/science.1221711
Loss of the tumor suppressor BAP1 causes myeloid transformation
Abstract
De-ubiquitinating enzyme BAP1 is mutated in a hereditary cancer syndrome with increased risk of mesothelioma and uveal melanoma. Somatic BAP1 mutations occur in various malignancies. We show that mouse Bap1 gene deletion is lethal during embryogenesis, but systemic or hematopoietic-restricted deletion in adults recapitulates features of human myelodysplastic syndrome (MDS). Knockin mice expressing BAP1 with a 3xFlag tag revealed that BAP1 interacts with host cell factor-1 (HCF-1), O-linked N-acetylglucosamine transferase (OGT), and the polycomb group proteins ASXL1 and ASXL2 in vivo. OGT and HCF-1 levels were decreased by Bap1 deletion, indicating a critical role for BAP1 in stabilizing these epigenetic regulators. Human ASXL1 is mutated frequently in chronic myelomonocytic leukemia (CMML) so an ASXL/BAP1 complex may suppress CMML. A BAP1 catalytic mutation found in a MDS patient implies that BAP1 loss of function has similar consequences in mice and humans.
Figures
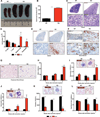


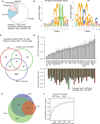
Comment in
-
Cancer. Emerging anatomy of the BAP1 tumor suppressor system.Science. 2012 Sep 21;337(6101):1463-4. doi: 10.1126/science.1228463. Science. 2012. PMID: 22997315 No abstract available.
References
-
- Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097. - PubMed
-
- Jensen DE, Rauscher FJ., 3rd Defining biochemical functions for the BRCA1 tumor suppressor protein: analysis of the BRCA1 binding protein BAP1. Cancer Lett. 1999;(143 Suppl 1):S13. - PubMed
-
- Guo G, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

