Expression of the RET proto-oncogene is regulated by TFAP2C in breast cancer independent of the estrogen receptor
- PMID: 22878616
- PMCID: PMC3697477
- DOI: 10.1245/s10434-012-2570-5
Expression of the RET proto-oncogene is regulated by TFAP2C in breast cancer independent of the estrogen receptor
Abstract
Background: The RET proto-oncogene is expressed as part of the estrogen receptor (ER) cluster in breast cancer. We sought to determine if TFAP2C regulates Ret expression directly or indirectly through ER.
Methods: Chromatin immunoprecipitation sequencing (ChIP-Seq) and gel-shift assay were used to identify TFAP2C binding sites in the RET promoter in four breast cancer cell lines. Ret mRNA and protein levels were evaluated in ER-positive and ER-negative breast cancer cell lines after knockdown of TFAP2C. Luciferase expression assay was performed to assess expression from two of the identified sites.
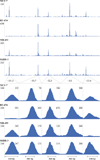
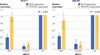
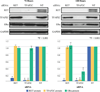
Results: ChIP-Seq identified five main binding peaks for TFAP2C in the RET promoter at -101.5 kb, -50.7 kb, -32.5 kb, +5.0 kb, and +33.6 from the RET transcriptional start site. Binding at three of the AP-2 sites was conserved across all four cell lines, whereas the RET -101.5 and RET +33.6 sites were each found to be unbound by TFAP2C in one cell line. A TFAP2C consensus element was confirmed for all five sites. Knockdown of TFAP2C by siRNA in ER-positive MCF-7 cells resulted in significant down regulation of Ret mRNA compared to nontargeting (NT) siRNA (0.09 vs. 1.0, P < 0.001). Knockdown of TFAP2C in ER-negative MDA-MB-453 cells also led to a significant reduction in Ret mRNA compared to NT siRNA (0.16 vs. 1.0, P < 0.001). In MCF-7 cells, knockdown of TFAP2C abrogated Ret protein expression (0.02 vs. 1.0, P < 0.001) before reduction in ER.
Conclusions: TFAP2C regulates expression of the RET proto-oncogene through five AP-2 regulatory sites in the RET promoter. Regulation of Ret by TFAP2C occurs independently of ER expression in breast carcinoma.
Figures

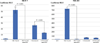
References
-
- Tozlu S, Girault I, Vacher S, et al. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription–PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. - PubMed
-
- Esseghir S, Todd SK, Hunt T, et al. A role for glial cell derived neurotrophic factor induced expression by inflammatory cytokines and RET/GFR alpha 1 receptor up-regulation in breast cancer. Cancer Res. 2007;67:11732–11741. - PubMed
-
- Boulay A, Breuleux M, Stephan C, et al. The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res. 2008;68:3743–3751. - PubMed
-
- Plaza-Menacho I, Morandi A, Robertson D, et al. Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010;29:4648–4657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

