Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci
- PMID: 22878891
- PMCID: PMC3887839
- DOI: 10.1007/s10059-012-0103-5
Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci
Abstract
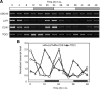
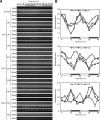
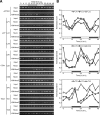
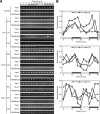
Circadian clock genes are regulated by a transcriptional-translational feedback loop. In Arabidopsis, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) transcripts are highly expressed in the morning. Translated LHY and CCA1 proteins repress the expression of TIMING OF CAB EXPRESSION 1 (TOC1), which peaks in the evening. TOC1 protein induces expression of LHY and CCA1, forming a negative feedback loop which is believed to constitute the oscillatory mechanism of the clock. The rhythmic oscillation of mouse clock genes mPERIOD 1 (mPER1) and mPER2 has been correlated with regular alteration of chromatin structure through histone acetylation/deacetylation. However, little is known about the relationship between the transcriptional activity of Arabidopsis clock genes and their chromatin status. Here, we report that histone H3 acetylation (H3Ac) and H3 lysine 4 tri-methylation (H3K4me3) levels at LHY, CCA1, and TOC1 are positively correlated with the rhythmic transcript levels of these genes, whereas H3K36me2 level shows a negative correlation. Thus, our study suggests rhythmic transcription of Arabidopsis clock genes might be regulated by rhythmic histone modification, and it provides a platform for future identification of clock-controlling histone modifiers.
Figures


Similar articles
-
Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms.Plant Cell Physiol. 2012 Dec;53(12):2016-29. doi: 10.1093/pcp/pcs148. Epub 2012 Nov 4. Plant Cell Physiol. 2012. PMID: 23128602 Free PMC article.
-
The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes.Nucleic Acids Res. 2018 Nov 16;46(20):10669-10681. doi: 10.1093/nar/gky749. Nucleic Acids Res. 2018. PMID: 30124938 Free PMC article.
-
Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation.Plant J. 2011 Apr;66(2):318-29. doi: 10.1111/j.1365-313X.2011.04484.x. Epub 2011 Feb 16. Plant J. 2011. PMID: 21205033
-
The circadian system of Arabidopsis thaliana: forward and reverse genetic approaches.Chronobiol Int. 1999 Jan;16(1):1-16. doi: 10.3109/07420529908998708. Chronobiol Int. 1999. PMID: 10023572 Review.
-
MYB transcription factors in the Arabidopsis circadian clock.J Exp Bot. 2002 Jul;53(374):1551-7. doi: 10.1093/jxb/erf027. J Exp Bot. 2002. PMID: 12096093 Review.
Cited by
-
The Plant Circadian Clock and Chromatin Modifications.Genes (Basel). 2018 Nov 20;9(11):561. doi: 10.3390/genes9110561. Genes (Basel). 2018. PMID: 30463332 Free PMC article. Review.
-
Circadian rhythms in stem cells and their therapeutic potential.Stem Cell Res Ther. 2025 Feb 23;16(1):85. doi: 10.1186/s13287-025-04178-9. Stem Cell Res Ther. 2025. PMID: 39988679 Free PMC article. Review.
-
The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis.Commun Biol. 2019 Apr 23;2:143. doi: 10.1038/s42003-019-0377-7. eCollection 2019. Commun Biol. 2019. PMID: 31044168 Free PMC article.
-
Integration of rhythmic metabolome and transcriptome provides insights into the transmission of rhythmic fluctuations and temporal diversity of metabolism in rice.Sci China Life Sci. 2022 Sep;65(9):1794-1810. doi: 10.1007/s11427-021-2064-7. Epub 2022 Mar 7. Sci China Life Sci. 2022. PMID: 35287184
-
Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice.Nucleic Acids Res. 2023 Sep 22;51(17):9001-9018. doi: 10.1093/nar/gkad658. Nucleic Acids Res. 2023. PMID: 37572350 Free PMC article.
References
-
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. - PubMed
-
- Belden W.J., Loros J.J., Dunlap J.C. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. - PubMed
-
- Brown S.A., Ripperger J., Kadener S., Fleury-Olela F., Vilbois F., Rosbash M., Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. - PubMed
-
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

