Identification and validation of potential conserved microRNAs and their targets in peach (Prunus persica)
- PMID: 22878892
- PMCID: PMC3887836
- DOI: 10.1007/s10059-012-0004-7
Identification and validation of potential conserved microRNAs and their targets in peach (Prunus persica)
Abstract
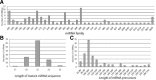
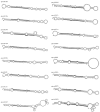
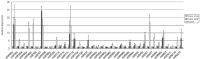
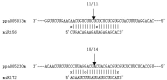
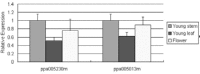
MicroRNAs are a class of small, endogenous, non-coding RNA molecules that negatively regulate gene expression at the transcriptional or the post-transcriptional level. Although a large number of miRNAs have been identified in many plant species, especially from model plants and crops, they remain largely unknown in peach. In this study, 110 potential miRNAs belonging to 37 families were identified using computational methods. A total of 43 potential targets were found for 21 families based on near-perfect or perfect complementarity between the plant miRNA and the target sequences. A majority of the targets were transcription factors which play important roles in peach development. qRT-PCR analysis of RNA samples prepared from different peach tissues for 25 miRNA families revealed that miRNAs were differentially expressed in different tissues. Furthermore, two target genes were experimentally verified by detection of the miRNA-mediated mRNA cleavage sites in peach using RNA ligase-mediated 5' rapid amplification of cDNA ends (RLM-RACE). Finally, we studied the expression pattern of the two target genes in three different tissues of peach to further understand the mechanism of the interaction between miRNAs and their target genes.
Figures
Similar articles
-
Identification of miRNAs and their target genes in peach (Prunus persica L.) using high-throughput sequencing and degradome analysis.PLoS One. 2013 Nov 13;8(11):e79090. doi: 10.1371/journal.pone.0079090. eCollection 2013. PLoS One. 2013. PMID: 24236092 Free PMC article.
-
Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs.BMC Plant Biol. 2012 Aug 21;12:149. doi: 10.1186/1471-2229-12-149. BMC Plant Biol. 2012. PMID: 22909020 Free PMC article.
-
Genome wide identification of chilling responsive microRNAs in Prunus persica.BMC Genomics. 2012 Sep 15;13:481. doi: 10.1186/1471-2164-13-481. BMC Genomics. 2012. PMID: 22978558 Free PMC article.
-
Plant microRNA: a small regulatory molecule with big impact.Dev Biol. 2006 Jan 1;289(1):3-16. doi: 10.1016/j.ydbio.2005.10.036. Epub 2005 Dec 1. Dev Biol. 2006. PMID: 16325172 Review.
-
MicroRNAs in fruit trees: discovery, diversity and future research directions.Plant Biol (Stuttg). 2014 Sep;16(5):856-65. doi: 10.1111/plb.12153. Epub 2014 Feb 19. Plant Biol (Stuttg). 2014. PMID: 24750383 Review.
Cited by
-
Prunus transcription factors: breeding perspectives.Front Plant Sci. 2015 Jun 12;6:443. doi: 10.3389/fpls.2015.00443. eCollection 2015. Front Plant Sci. 2015. PMID: 26124770 Free PMC article. Review.
-
Identification and characterization of Prunus persica miRNAs in response to UVB radiation in greenhouse through high-throughput sequencing.BMC Genomics. 2017 Dec 2;18(1):938. doi: 10.1186/s12864-017-4347-5. BMC Genomics. 2017. PMID: 29197334 Free PMC article.
-
Identification and characterisation of microRNAs and their target genes in phosphate-starved Nicotiana benthamiana by small RNA deep sequencing and 5'RACE analysis.BMC Genomics. 2018 Dec 17;19(1):940. doi: 10.1186/s12864-018-5258-9. BMC Genomics. 2018. PMID: 30558535 Free PMC article.
-
Identification of miRNAs and their target genes in peach (Prunus persica L.) using high-throughput sequencing and degradome analysis.PLoS One. 2013 Nov 13;8(11):e79090. doi: 10.1371/journal.pone.0079090. eCollection 2013. PLoS One. 2013. PMID: 24236092 Free PMC article.
-
Computational prediction of miRNAs and their targets in Phaseolus vulgaris using simple sequence repeat signatures.BMC Plant Biol. 2015 Jun 12;15:140. doi: 10.1186/s12870-015-0516-3. BMC Plant Biol. 2015. PMID: 26067253 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

