Discovery of multi-dimensional modules by integrative analysis of cancer genomic data
- PMID: 22879375
- PMCID: PMC3479191
- DOI: 10.1093/nar/gks725
Discovery of multi-dimensional modules by integrative analysis of cancer genomic data
Abstract
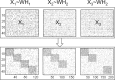
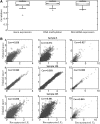
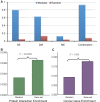
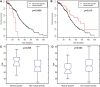
Recent technology has made it possible to simultaneously perform multi-platform genomic profiling (e.g. DNA methylation (DM) and gene expression (GE)) of biological samples, resulting in so-called 'multi-dimensional genomic data'. Such data provide unique opportunities to study the coordination between regulatory mechanisms on multiple levels. However, integrative analysis of multi-dimensional genomics data for the discovery of combinatorial patterns is currently lacking. Here, we adopt a joint matrix factorization technique to address this challenge. This method projects multiple types of genomic data onto a common coordinate system, in which heterogeneous variables weighted highly in the same projected direction form a multi-dimensional module (md-module). Genomic variables in such modules are characterized by significant correlations and likely functional associations. We applied this method to the DM, GE, and microRNA expression data of 385 ovarian cancer samples from the The Cancer Genome Atlas project. These md-modules revealed perturbed pathways that would have been overlooked with only a single type of data, uncovered associations between different layers of cellular activities and allowed the identification of clinically distinct patient subgroups. Our study provides an useful protocol for uncovering hidden patterns and their biological implications in multi-dimensional 'omic' data.
Figures


References
-
- Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. - PubMed
-
- Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, et al. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000;24:236–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

