Fate-restricted neural progenitors in the mammalian cerebral cortex
- PMID: 22879516
- PMCID: PMC4287277
- DOI: 10.1126/science.1223616
Fate-restricted neural progenitors in the mammalian cerebral cortex
Abstract
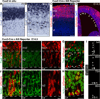
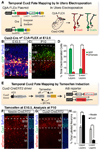
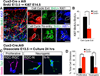
During development of the mammalian cerebral cortex, radial glial cells (RGCs) generate layer-specific subtypes of excitatory neurons in a defined temporal sequence, in which lower-layer neurons are formed before upper-layer neurons. It has been proposed that neuronal subtype fate is determined by birthdate through progressive restriction of the neurogenic potential of a common RGC progenitor. Here, we demonstrate that the murine cerebral cortex contains RGC sublineages with distinct fate potentials. Using in vivo genetic fate mapping and in vitro clonal analysis, we identified an RGC lineage that is intrinsically specified to generate only upper-layer neurons, independently of niche and birthdate. Because upper cortical layers were expanded during primate evolution, amplification of this RGC pool may have facilitated human brain evolution.
Figures

References
-
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. - PubMed
-
- Hevner RF, et al. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. - PubMed
-
- Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers IIIV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

