Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein
- PMID: 22879595
- PMCID: PMC3460473
- DOI: 10.1074/jbc.M112.393132
Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein
Abstract
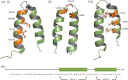
Many viruses encode scaffolding and coat proteins that co-assemble to form procapsids, which are transient precursor structures leading to progeny virions. In bacteriophage P22, the association of scaffolding and coat proteins is mediated mainly by ionic interactions. The coat protein-binding domain of scaffolding protein is a helix turn helix structure near the C terminus with a high number of charged surface residues. Residues Arg-293 and Lys-296 are particularly important for coat protein binding. The two helices contact each other through hydrophobic side chains. In this study, substitution of the residues of the interface between the helices, and the residues in the β-turn, by aspartic acid was used examine the importance of the conformation of the domain in coat binding. These replacements strongly affected the ability of the scaffolding protein to interact with coat protein. The severity of the defect in the association of scaffolding protein to coat protein was dependent on location, with substitutions at residues in the turn and helix 2 causing the most significant effects. Substituting aspartic acid for hydrophobic interface residues dramatically perturbs the stability of the structure, but similar substitutions in the turn had much less effect on the integrity of this domain, as determined by circular dichroism. We propose that the binding of scaffolding protein to coat protein is dependent on angle of the β-turn and the orientation of the charged surface on helix 2. Surprisingly, formation of the highly complex procapsid structure depends on a relatively simple interaction.
Figures






Similar articles
-
Decoding bacteriophage P22 assembly: identification of two charged residues in scaffolding protein responsible for coat protein interaction.Virology. 2011 Dec 5;421(1):1-11. doi: 10.1016/j.virol.2011.09.005. Epub 2011 Oct 4. Virology. 2011. PMID: 21974803 Free PMC article.
-
Highly specific salt bridges govern bacteriophage P22 icosahedral capsid assembly: identification of the site in coat protein responsible for interaction with scaffolding protein.J Virol. 2014 May;88(10):5287-97. doi: 10.1128/JVI.00036-14. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600011 Free PMC article.
-
Electrostatic interactions drive scaffolding/coat protein binding and procapsid maturation in bacteriophage P22.Virology. 1998 Oct 25;250(2):337-49. doi: 10.1006/viro.1998.9386. Virology. 1998. PMID: 9792844
-
A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly.J Virol. 2015 Oct;89(20):10569-79. doi: 10.1128/JVI.01629-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269173 Free PMC article.
-
Virus particle maturation: insights into elegantly programmed nanomachines.Curr Opin Struct Biol. 2010 Apr;20(2):210-6. doi: 10.1016/j.sbi.2010.01.004. Epub 2010 Feb 9. Curr Opin Struct Biol. 2010. PMID: 20149636 Free PMC article. Review.
Cited by
-
Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor.Virology. 2013 Jun 5;440(2):117-33. doi: 10.1016/j.virol.2013.02.023. Epub 2013 Apr 4. Virology. 2013. PMID: 23562538 Free PMC article.
-
Effects of an early conformational switch defect during ϕX174 morphogenesis are belatedly manifested late in the assembly pathway.J Virol. 2013 Mar;87(5):2518-25. doi: 10.1128/JVI.02839-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255785 Free PMC article.
-
Structural Model of Bacteriophage P22 Scaffolding Protein in a Procapsid by Magic-Angle Spinning NMR.bioRxiv [Preprint]. 2024 Nov 1:2024.11.01.621488. doi: 10.1101/2024.11.01.621488. bioRxiv. 2024. PMID: 39554170 Free PMC article. Preprint.
-
Phi29 assembly intermediates reveal how scaffold interactions with capsid protein drive capsid construction and maturation.Sci Adv. 2025 Mar 21;11(12):eadk8779. doi: 10.1126/sciadv.adk8779. Epub 2025 Mar 19. Sci Adv. 2025. PMID: 40106547 Free PMC article.
-
Bacteriophage P22 SieA-mediated superinfection exclusion.mBio. 2024 Feb 14;15(2):e0216923. doi: 10.1128/mbio.02169-23. Epub 2024 Jan 18. mBio. 2024. PMID: 38236051 Free PMC article.
References
-
- King J., Casjens S. (1974) Catalytic head assembling protein in virus morphogenesis. Nature 251, 112–119 - PubMed
-
- Prevelige P. E., Fane B. A. (2012) Building the machines. Scaffolding protein functions during bacteriophage morphogenesis. Adv. Exp. Med. Biol. 726, 325–350 - PubMed
-
- Rishovd S., Marvik O. J., Jacobsen E., Lindqvist B. H. (1994) Bacteriophage P2 and P4 morphogenesis. Identification and characterization of the portal protein. Virology 200, 744–751 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

