Complete mitochondrial genome of the free-living earwig, Challia fletcheri (Dermaptera: Pygidicranidae) and phylogeny of Polyneoptera
- PMID: 22879905
- PMCID: PMC3412835
- DOI: 10.1371/journal.pone.0042056
Complete mitochondrial genome of the free-living earwig, Challia fletcheri (Dermaptera: Pygidicranidae) and phylogeny of Polyneoptera
Abstract
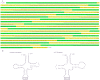
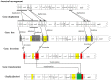
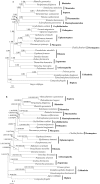
The insect order Dermaptera, belonging to Polyneoptera, includes ∼2,000 extant species, but no dermapteran mitochondrial genome has been sequenced. We sequenced the complete mitochondrial genome of the free-living earwig, Challia fletcheri, compared its genomic features to other available mitochondrial sequences from polyneopterous insects. In addition, the Dermaptera, together with the other known polyneopteran mitochondrial genome sequences (protein coding, ribosomal RNA, and transfer RNA genes), were employed to understand the phylogeny of Polyneoptera, one of the least resolved insect phylogenies, with emphasis on the placement of Dermaptera. The complete mitochondrial genome of C. fletcheri presents the following several unusual features: the longest size in insects is 20,456 bp; it harbors the largest tandem repeat units (TRU) among insects; it displays T- and G-skewness on the major strand and A- and C-skewness on the minor strand, which is a reversal of the general pattern found in most insect mitochondrial genomes, and it possesses a unique gene arrangement characterized by a series of gene translocations and/or inversions. The reversal pattern of skewness is explained in terms of inversion of replication origin. All phylogenetic analyses consistently placed Dermaptera as the sister to Plecoptera, leaving them as the most basal lineage of Polyneoptera or sister to Ephemeroptera, and placed Odonata consistently as the most basal lineage of the Pterygota.
Conflict of interest statement
Figures




References
-
- Brown WM (1985) The mitochondrial genome of animals. In: Macintyre RJ, editor. Molecular Evolution Genetics. New York: Plenum.
-
- Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141: 173–216. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

