Structure determination and functional analysis of a chromate reductase from Gluconacetobacter hansenii
- PMID: 22879982
- PMCID: PMC3412864
- DOI: 10.1371/journal.pone.0042432
Structure determination and functional analysis of a chromate reductase from Gluconacetobacter hansenii
Abstract
Environmental protection through biological mechanisms that aid in the reductive immobilization of toxic metals (e.g., chromate and uranyl) has been identified to involve specific NADH-dependent flavoproteins that promote cell viability. To understand the enzyme mechanisms responsible for metal reduction, the enzyme kinetics of a putative chromate reductase from Gluconacetobacter hansenii (Gh-ChrR) was measured and the crystal structure of the protein determined at 2.25 Å resolution. Gh-ChrR catalyzes the NADH-dependent reduction of chromate, ferricyanide, and uranyl anions under aerobic conditions. Kinetic measurements indicate that NADH acts as a substrate inhibitor; catalysis requires chromate binding prior to NADH association. The crystal structure of Gh-ChrR shows the protein is a homotetramer with one bound flavin mononucleotide (FMN) per subunit. A bound anion is visualized proximal to the FMN at the interface between adjacent subunits within a cationic pocket, which is positioned at an optimal distance for hydride transfer. Site-directed substitutions of residues proposed to involve in both NADH and metal anion binding (N85A or R101A) result in 90-95% reductions in enzyme efficiencies for NADH-dependent chromate reduction. In comparison site-directed substitution of a residue (S118A) participating in the coordination of FMN in the active site results in only modest (50%) reductions in catalytic efficiencies, consistent with the presence of a multitude of side chains that position the FMN in the active site. The proposed proximity relationships between metal anion binding site and enzyme cofactors is discussed in terms of rational design principles for the use of enzymes in chromate and uranyl bioremediation.
Conflict of interest statement
Figures
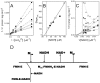



References
-
- Wall JD, Krumholz LR (2006) Uranium reduction. Annu Rev Microbiol 60: 149–166. - PubMed
-
- Fredrickson JK, Zachara JM (2008) Electron transfer at the microbe-mineral interface: a grand challenge in biogeochemistry. Geobiology 6: 245–253. - PubMed
-
- Lovley DR, Phillips EJP (2002) Bioremediation of uranium contamination with enzymatic uranium reduction. Environmental Science & Technology 26: 2228–2234.
-
- Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, et al. (2005) Extracellular electron transfer via microbial nanowires. Nature 435: 1098–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

