Detection of bladder cancer using proteomic profiling of urine sediments
- PMID: 22879988
- PMCID: PMC3411788
- DOI: 10.1371/journal.pone.0042452
Detection of bladder cancer using proteomic profiling of urine sediments
Abstract
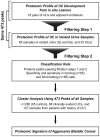
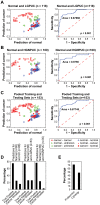
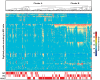
We used protein expression profiles to develop a classification rule for the detection and prognostic assessment of bladder cancer in voided urine samples. Using the Ciphergen PBS II ProteinChip Reader, we analyzed the protein profiles of 18 pairs of samples of bladder tumor and adjacent urothelium tissue, a training set of 85 voided urine samples (32 controls and 53 bladder cancer), and a blinded testing set of 68 voided urine samples (33 controls and 35 bladder cancer). Using t-tests, we identified 473 peaks showing significant differential expression across different categories of paired bladder tumor and adjacent urothelial samples compared to normal urothelium. Then the intensities of those 473 peaks were examined in a training set of voided urine samples. Using this approach, we identified 41 protein peaks that were differentially expressed in both sets of samples. The expression pattern of the 41 protein peaks was used to classify the voided urine samples as malignant or benign. This approach yielded a sensitivity and specificity of 59% and 90%, respectively, on the training set and 80% and 100%, respectively, on the testing set. The proteomic classification rule performed with similar accuracy in low- and high-grade bladder carcinomas. In addition, we used hierarchical clustering with all 473 protein peaks on 65 benign voided urine samples, 88 samples from patients with clinically evident bladder cancer, and 127 samples from patients with a history of bladder cancer to classify the samples into Cluster A or B. The tumors in Cluster B were characterized by clinically aggressive behavior with significantly shorter metastasis-free and disease-specific survival.
Conflict of interest statement
Figures




References
-
- Dinney CP, McConkey DJ, Millikan RE, Wu X, Bar-Eli M, et al. (2004) Focus on bladder cancer. Cancer Cell 6: 111–116. - PubMed
-
- Spiess PE, Czerniak B (2006) Dual-track pathway of bladder carcinogenesis: practical implications. Arch Pathol Lab Med 130: 844–852. - PubMed
-
- Gazdar AF, Czerniak B (2001) Filling the void: urinary markers for bladder cancer risk and diagnosis. J Natl Cancer Inst 93: 413–415. - PubMed
-
- Rogers MA, Clarke P, Noble J, Munro NP, Paul A, et al. (2003) Proteomic profiling of urinary proteins in renal cancer by surface enhanced laser desorption ionization and neural-network analysis: identification of key issues affecting potential clinical utility. Cancer Res 63: 6971–6983. - PubMed
-
- Schaub S, Wilkins J, Weiler T, Sangster K, Rush D, et al. (2004) Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int 65: 323–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

