Structural and functional characterization of ribosomal protein gene introns in sponges
- PMID: 22880015
- PMCID: PMC3412847
- DOI: 10.1371/journal.pone.0042523
Structural and functional characterization of ribosomal protein gene introns in sponges
Abstract
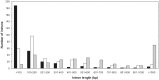
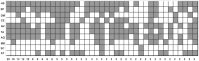
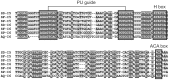
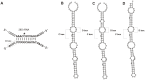
Ribosomal protein genes (RPGs) are a powerful tool for studying intron evolution. They exist in all three domains of life and are much conserved. Accumulating genomic data suggest that RPG introns in many organisms abound with non-protein-coding-RNAs (ncRNAs). These ancient ncRNAs are small nucleolar RNAs (snoRNAs) essential for ribosome assembly. They are also mobile genetic elements and therefore probably important in diversification and enrichment of transcriptomes through various mechanisms such as intron/exon gain/loss. snoRNAs in basal metazoans are poorly characterized. We examined 449 RPG introns, in total, from four demosponges: Amphimedon queenslandica, Suberites domuncula, Suberites ficus and Suberites pagurorum and showed that RPG introns from A. queenslandica share position conservancy and some structural similarity with "higher" metazoans. Moreover, our study indicates that mobile element insertions play an important role in the evolution of their size. In four sponges 51 snoRNAs were identified. The analysis showed discrepancies between the snoRNA pools of orthologous RPG introns between S. domuncula and A. queenslandica. Furthermore, these two sponges show as much conservancy of RPG intron positions between each other as between themselves and human. Sponges from the Suberites genus show consistency in RPG intron position conservation. However, significant differences in some of the orthologous RPG introns of closely related sponges were observed. This indicates that RPG introns are dynamic even on these shorter evolutionary time scales.
Conflict of interest statement
Figures
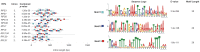


Similar articles
-
Conservation of the positions of metazoan introns from sponges to humans.Gene. 2002 Aug 7;295(2):299-309. doi: 10.1016/s0378-1119(02)00690-x. Gene. 2002. PMID: 12354665
-
Src proteins/src genes: from sponges to mammals.Gene. 2004 Nov 24;342(2):251-61. doi: 10.1016/j.gene.2004.07.044. Gene. 2004. PMID: 15527984
-
Complete genomic structure of human rpS3: identification of functional U15b snoRNA in the fifth intron.Gene. 2002 Mar 20;286(2):291-7. doi: 10.1016/s0378-1119(02)00502-4. Gene. 2002. PMID: 11943484
-
Novel intron-encoded small nucleolar RNAs with long sequence complementarities to mature rRNAs involved in ribosome biogenesis.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):835-43. doi: 10.1139/o95-091. Biochem Cell Biol. 1995. PMID: 8721999 Review.
-
Approaches for a sustainable use of the bioactive potential in sponges: analysis of gene clusters, differential display of mRNA and DNA chips.Prog Mol Subcell Biol. 2003;37:199-230. doi: 10.1007/978-3-642-55519-0_8. Prog Mol Subcell Biol. 2003. PMID: 15825645 Review.
Cited by
-
Functional and Structural Characterization of FAU Gene/Protein from Marine Sponge Suberites domuncula.Mar Drugs. 2015 Jul 7;13(7):4179-96. doi: 10.3390/md13074179. Mar Drugs. 2015. PMID: 26198235 Free PMC article.
-
Sponges: A Reservoir of Genes Implicated in Human Cancer.Mar Drugs. 2018 Jan 10;16(1):20. doi: 10.3390/md16010020. Mar Drugs. 2018. PMID: 29320389 Free PMC article. Review.
References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920. - PubMed
-
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. - PubMed
-
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102: 615–623. - PubMed
-
- Kressler D, Hurt E, Bassler J (2009) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

